
Beihai weivirus-like virus 15
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.42
Get precalculated fractions of proteins
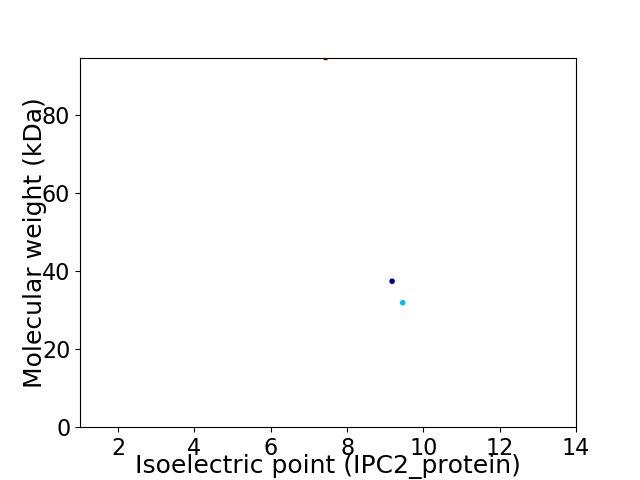
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLC4|A0A1L3KLC4_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 15 OX=1922743 PE=4 SV=1
MM1 pKa = 6.9FTRR4 pKa = 11.84FARR7 pKa = 11.84DD8 pKa = 3.08GSIRR12 pKa = 11.84RR13 pKa = 11.84VCAFCDD19 pKa = 3.54RR20 pKa = 11.84GALDD24 pKa = 4.81AVADD28 pKa = 4.41LDD30 pKa = 4.6GDD32 pKa = 3.9LCLEE36 pKa = 3.97YY37 pKa = 10.66DD38 pKa = 3.88VWPDD42 pKa = 3.48CQRR45 pKa = 11.84CAVAYY50 pKa = 7.37WSPFACCHH58 pKa = 6.3PPRR61 pKa = 11.84GPARR65 pKa = 11.84RR66 pKa = 11.84ILHH69 pKa = 6.26LAVEE73 pKa = 4.39DD74 pKa = 4.51AEE76 pKa = 5.13AILYY80 pKa = 9.91NYY82 pKa = 8.92TSASDD87 pKa = 3.66ASLTATLVNRR97 pKa = 11.84LRR99 pKa = 11.84GFGCPQVSVALLRR112 pKa = 11.84LLPPILDD119 pKa = 5.21AIQADD124 pKa = 4.23RR125 pKa = 11.84PYY127 pKa = 11.09RR128 pKa = 11.84DD129 pKa = 4.02ALLDD133 pKa = 3.44MRR135 pKa = 11.84KK136 pKa = 9.57EE137 pKa = 4.06DD138 pKa = 3.71GAGVEE143 pKa = 4.42SAQRR147 pKa = 11.84TSCCEE152 pKa = 3.86GLCDD156 pKa = 5.01GAQQRR161 pKa = 11.84PPRR164 pKa = 11.84WWFRR168 pKa = 11.84RR169 pKa = 11.84PARR172 pKa = 11.84VGQPADD178 pKa = 4.2PGPPGPNGGGGVGVGGPEE196 pKa = 3.59QRR198 pKa = 11.84ANTTAANGTPNSSTTTAGPIRR219 pKa = 11.84APGGARR225 pKa = 11.84EE226 pKa = 4.59GGRR229 pKa = 11.84ADD231 pKa = 4.28PGPGRR236 pKa = 11.84AGGGTPPATGTSHH249 pKa = 6.98APAGEE254 pKa = 4.32GARR257 pKa = 11.84STEE260 pKa = 4.09QQQSGPARR268 pKa = 11.84PRR270 pKa = 11.84LCPPPGAGSLRR281 pKa = 11.84QQDD284 pKa = 3.13VDD286 pKa = 4.0RR287 pKa = 11.84GVGTDD292 pKa = 3.41TTAPPPGRR300 pKa = 11.84IDD302 pKa = 3.56PPHH305 pKa = 6.5AVGAILRR312 pKa = 11.84SEE314 pKa = 4.1AHH316 pKa = 6.93PARR319 pKa = 11.84CEE321 pKa = 3.97TQVTVEE327 pKa = 3.85RR328 pKa = 11.84LEE330 pKa = 4.19PPKK333 pKa = 10.63EE334 pKa = 3.49RR335 pKa = 11.84VAISRR340 pKa = 11.84ATNPCDD346 pKa = 3.27PQHH349 pKa = 6.72FDD351 pKa = 3.21EE352 pKa = 5.32SLTNVMAATAQRR364 pKa = 11.84VEE366 pKa = 4.22RR367 pKa = 11.84YY368 pKa = 8.34QPKK371 pKa = 10.0FKK373 pKa = 10.5PGPQLRR379 pKa = 11.84ARR381 pKa = 11.84LEE383 pKa = 3.96KK384 pKa = 10.9VRR386 pKa = 11.84DD387 pKa = 3.89SIIEE391 pKa = 3.94RR392 pKa = 11.84VLTKK396 pKa = 10.61KK397 pKa = 10.52KK398 pKa = 9.75IYY400 pKa = 9.67QWLDD404 pKa = 3.03EE405 pKa = 4.52HH406 pKa = 6.34PTLRR410 pKa = 11.84SAMSKK415 pKa = 9.79AWSDD419 pKa = 3.2EE420 pKa = 3.91RR421 pKa = 11.84AQRR424 pKa = 11.84AVEE427 pKa = 3.99TLRR430 pKa = 11.84GLGFDD435 pKa = 3.62VRR437 pKa = 11.84ITHH440 pKa = 6.14EE441 pKa = 4.07ASIKK445 pKa = 11.08DD446 pKa = 3.45EE447 pKa = 4.0VLPNRR452 pKa = 11.84GKK454 pKa = 10.61APRR457 pKa = 11.84LIISCGDD464 pKa = 3.36QGQLCALIHH473 pKa = 5.06VACFEE478 pKa = 4.05GLLYY482 pKa = 10.99DD483 pKa = 3.81HH484 pKa = 7.03FEE486 pKa = 3.99KK487 pKa = 10.77RR488 pKa = 11.84SIKK491 pKa = 10.16HH492 pKa = 5.35RR493 pKa = 11.84AKK495 pKa = 10.65ADD497 pKa = 3.4ALGEE501 pKa = 3.98VSGRR505 pKa = 11.84AKK507 pKa = 10.41QEE509 pKa = 3.51EE510 pKa = 5.07GFVKK514 pKa = 10.36RR515 pKa = 11.84YY516 pKa = 10.42LEE518 pKa = 4.44GDD520 pKa = 3.53GKK522 pKa = 10.96AWDD525 pKa = 3.72RR526 pKa = 11.84TITPRR531 pKa = 11.84LRR533 pKa = 11.84EE534 pKa = 3.99LLEE537 pKa = 4.35NPLLEE542 pKa = 4.68HH543 pKa = 6.31VANTIYY549 pKa = 10.94ARR551 pKa = 11.84WDD553 pKa = 3.98AIGSLSYY560 pKa = 9.34WASVDD565 pKa = 2.88IKK567 pKa = 10.96DD568 pKa = 3.36RR569 pKa = 11.84KK570 pKa = 9.87KK571 pKa = 9.62PIRR574 pKa = 11.84RR575 pKa = 11.84LVYY578 pKa = 9.59RR579 pKa = 11.84KK580 pKa = 10.0GSRR583 pKa = 11.84AHH585 pKa = 6.04MFLEE589 pKa = 4.54AFRR592 pKa = 11.84ASGDD596 pKa = 3.46RR597 pKa = 11.84GTSCLNWVVNYY608 pKa = 10.2SCWLAVLVAYY618 pKa = 10.08EE619 pKa = 4.72DD620 pKa = 3.26IDD622 pKa = 4.28RR623 pKa = 11.84AVTQPNWRR631 pKa = 11.84WFRR634 pKa = 11.84PAPHH638 pKa = 6.72YY639 pKa = 10.57LRR641 pKa = 11.84NDD643 pKa = 2.59KK644 pKa = 9.73RR645 pKa = 11.84TLSTRR650 pKa = 11.84RR651 pKa = 11.84KK652 pKa = 8.02MFMDD656 pKa = 3.94SVYY659 pKa = 10.81EE660 pKa = 4.26GDD662 pKa = 4.44DD663 pKa = 3.71SFVMISTDD671 pKa = 3.6RR672 pKa = 11.84PLEE675 pKa = 4.71DD676 pKa = 5.23IEE678 pKa = 4.94AAWASLGFEE687 pKa = 3.92MKK689 pKa = 10.49LFEE692 pKa = 4.58RR693 pKa = 11.84RR694 pKa = 11.84PGSVVTFVGNEE705 pKa = 3.94CLVKK709 pKa = 10.91EE710 pKa = 4.58EE711 pKa = 4.49GLSDD715 pKa = 3.6LWLPQIARR723 pKa = 11.84NLTAAAEE730 pKa = 4.01QRR732 pKa = 11.84DD733 pKa = 4.48GGGDD737 pKa = 3.49PHH739 pKa = 6.37QHH741 pKa = 5.98SLACAVRR748 pKa = 11.84AQGYY752 pKa = 9.03KK753 pKa = 10.17RR754 pKa = 11.84YY755 pKa = 9.14PPLALHH761 pKa = 6.15YY762 pKa = 10.31LAIAKK767 pKa = 9.54SCRR770 pKa = 11.84DD771 pKa = 3.92SNTLAQPVAKK781 pKa = 8.97ATQYY785 pKa = 11.79AWFGNYY791 pKa = 9.89NPDD794 pKa = 3.13RR795 pKa = 11.84RR796 pKa = 11.84LEE798 pKa = 4.12PEE800 pKa = 4.37VYY802 pKa = 9.87EE803 pKa = 4.49LPAVDD808 pKa = 3.73YY809 pKa = 11.1HH810 pKa = 7.13PDD812 pKa = 3.4AEE814 pKa = 4.48ALVAAATGEE823 pKa = 4.14APDD826 pKa = 4.23RR827 pKa = 11.84NAVDD831 pKa = 3.96ALMTGSLAAEE841 pKa = 4.05WDD843 pKa = 3.72KK844 pKa = 10.91EE845 pKa = 4.33TLRR848 pKa = 11.84RR849 pKa = 11.84ILPRR853 pKa = 11.84SWIEE857 pKa = 3.56
MM1 pKa = 6.9FTRR4 pKa = 11.84FARR7 pKa = 11.84DD8 pKa = 3.08GSIRR12 pKa = 11.84RR13 pKa = 11.84VCAFCDD19 pKa = 3.54RR20 pKa = 11.84GALDD24 pKa = 4.81AVADD28 pKa = 4.41LDD30 pKa = 4.6GDD32 pKa = 3.9LCLEE36 pKa = 3.97YY37 pKa = 10.66DD38 pKa = 3.88VWPDD42 pKa = 3.48CQRR45 pKa = 11.84CAVAYY50 pKa = 7.37WSPFACCHH58 pKa = 6.3PPRR61 pKa = 11.84GPARR65 pKa = 11.84RR66 pKa = 11.84ILHH69 pKa = 6.26LAVEE73 pKa = 4.39DD74 pKa = 4.51AEE76 pKa = 5.13AILYY80 pKa = 9.91NYY82 pKa = 8.92TSASDD87 pKa = 3.66ASLTATLVNRR97 pKa = 11.84LRR99 pKa = 11.84GFGCPQVSVALLRR112 pKa = 11.84LLPPILDD119 pKa = 5.21AIQADD124 pKa = 4.23RR125 pKa = 11.84PYY127 pKa = 11.09RR128 pKa = 11.84DD129 pKa = 4.02ALLDD133 pKa = 3.44MRR135 pKa = 11.84KK136 pKa = 9.57EE137 pKa = 4.06DD138 pKa = 3.71GAGVEE143 pKa = 4.42SAQRR147 pKa = 11.84TSCCEE152 pKa = 3.86GLCDD156 pKa = 5.01GAQQRR161 pKa = 11.84PPRR164 pKa = 11.84WWFRR168 pKa = 11.84RR169 pKa = 11.84PARR172 pKa = 11.84VGQPADD178 pKa = 4.2PGPPGPNGGGGVGVGGPEE196 pKa = 3.59QRR198 pKa = 11.84ANTTAANGTPNSSTTTAGPIRR219 pKa = 11.84APGGARR225 pKa = 11.84EE226 pKa = 4.59GGRR229 pKa = 11.84ADD231 pKa = 4.28PGPGRR236 pKa = 11.84AGGGTPPATGTSHH249 pKa = 6.98APAGEE254 pKa = 4.32GARR257 pKa = 11.84STEE260 pKa = 4.09QQQSGPARR268 pKa = 11.84PRR270 pKa = 11.84LCPPPGAGSLRR281 pKa = 11.84QQDD284 pKa = 3.13VDD286 pKa = 4.0RR287 pKa = 11.84GVGTDD292 pKa = 3.41TTAPPPGRR300 pKa = 11.84IDD302 pKa = 3.56PPHH305 pKa = 6.5AVGAILRR312 pKa = 11.84SEE314 pKa = 4.1AHH316 pKa = 6.93PARR319 pKa = 11.84CEE321 pKa = 3.97TQVTVEE327 pKa = 3.85RR328 pKa = 11.84LEE330 pKa = 4.19PPKK333 pKa = 10.63EE334 pKa = 3.49RR335 pKa = 11.84VAISRR340 pKa = 11.84ATNPCDD346 pKa = 3.27PQHH349 pKa = 6.72FDD351 pKa = 3.21EE352 pKa = 5.32SLTNVMAATAQRR364 pKa = 11.84VEE366 pKa = 4.22RR367 pKa = 11.84YY368 pKa = 8.34QPKK371 pKa = 10.0FKK373 pKa = 10.5PGPQLRR379 pKa = 11.84ARR381 pKa = 11.84LEE383 pKa = 3.96KK384 pKa = 10.9VRR386 pKa = 11.84DD387 pKa = 3.89SIIEE391 pKa = 3.94RR392 pKa = 11.84VLTKK396 pKa = 10.61KK397 pKa = 10.52KK398 pKa = 9.75IYY400 pKa = 9.67QWLDD404 pKa = 3.03EE405 pKa = 4.52HH406 pKa = 6.34PTLRR410 pKa = 11.84SAMSKK415 pKa = 9.79AWSDD419 pKa = 3.2EE420 pKa = 3.91RR421 pKa = 11.84AQRR424 pKa = 11.84AVEE427 pKa = 3.99TLRR430 pKa = 11.84GLGFDD435 pKa = 3.62VRR437 pKa = 11.84ITHH440 pKa = 6.14EE441 pKa = 4.07ASIKK445 pKa = 11.08DD446 pKa = 3.45EE447 pKa = 4.0VLPNRR452 pKa = 11.84GKK454 pKa = 10.61APRR457 pKa = 11.84LIISCGDD464 pKa = 3.36QGQLCALIHH473 pKa = 5.06VACFEE478 pKa = 4.05GLLYY482 pKa = 10.99DD483 pKa = 3.81HH484 pKa = 7.03FEE486 pKa = 3.99KK487 pKa = 10.77RR488 pKa = 11.84SIKK491 pKa = 10.16HH492 pKa = 5.35RR493 pKa = 11.84AKK495 pKa = 10.65ADD497 pKa = 3.4ALGEE501 pKa = 3.98VSGRR505 pKa = 11.84AKK507 pKa = 10.41QEE509 pKa = 3.51EE510 pKa = 5.07GFVKK514 pKa = 10.36RR515 pKa = 11.84YY516 pKa = 10.42LEE518 pKa = 4.44GDD520 pKa = 3.53GKK522 pKa = 10.96AWDD525 pKa = 3.72RR526 pKa = 11.84TITPRR531 pKa = 11.84LRR533 pKa = 11.84EE534 pKa = 3.99LLEE537 pKa = 4.35NPLLEE542 pKa = 4.68HH543 pKa = 6.31VANTIYY549 pKa = 10.94ARR551 pKa = 11.84WDD553 pKa = 3.98AIGSLSYY560 pKa = 9.34WASVDD565 pKa = 2.88IKK567 pKa = 10.96DD568 pKa = 3.36RR569 pKa = 11.84KK570 pKa = 9.87KK571 pKa = 9.62PIRR574 pKa = 11.84RR575 pKa = 11.84LVYY578 pKa = 9.59RR579 pKa = 11.84KK580 pKa = 10.0GSRR583 pKa = 11.84AHH585 pKa = 6.04MFLEE589 pKa = 4.54AFRR592 pKa = 11.84ASGDD596 pKa = 3.46RR597 pKa = 11.84GTSCLNWVVNYY608 pKa = 10.2SCWLAVLVAYY618 pKa = 10.08EE619 pKa = 4.72DD620 pKa = 3.26IDD622 pKa = 4.28RR623 pKa = 11.84AVTQPNWRR631 pKa = 11.84WFRR634 pKa = 11.84PAPHH638 pKa = 6.72YY639 pKa = 10.57LRR641 pKa = 11.84NDD643 pKa = 2.59KK644 pKa = 9.73RR645 pKa = 11.84TLSTRR650 pKa = 11.84RR651 pKa = 11.84KK652 pKa = 8.02MFMDD656 pKa = 3.94SVYY659 pKa = 10.81EE660 pKa = 4.26GDD662 pKa = 4.44DD663 pKa = 3.71SFVMISTDD671 pKa = 3.6RR672 pKa = 11.84PLEE675 pKa = 4.71DD676 pKa = 5.23IEE678 pKa = 4.94AAWASLGFEE687 pKa = 3.92MKK689 pKa = 10.49LFEE692 pKa = 4.58RR693 pKa = 11.84RR694 pKa = 11.84PGSVVTFVGNEE705 pKa = 3.94CLVKK709 pKa = 10.91EE710 pKa = 4.58EE711 pKa = 4.49GLSDD715 pKa = 3.6LWLPQIARR723 pKa = 11.84NLTAAAEE730 pKa = 4.01QRR732 pKa = 11.84DD733 pKa = 4.48GGGDD737 pKa = 3.49PHH739 pKa = 6.37QHH741 pKa = 5.98SLACAVRR748 pKa = 11.84AQGYY752 pKa = 9.03KK753 pKa = 10.17RR754 pKa = 11.84YY755 pKa = 9.14PPLALHH761 pKa = 6.15YY762 pKa = 10.31LAIAKK767 pKa = 9.54SCRR770 pKa = 11.84DD771 pKa = 3.92SNTLAQPVAKK781 pKa = 8.97ATQYY785 pKa = 11.79AWFGNYY791 pKa = 9.89NPDD794 pKa = 3.13RR795 pKa = 11.84RR796 pKa = 11.84LEE798 pKa = 4.12PEE800 pKa = 4.37VYY802 pKa = 9.87EE803 pKa = 4.49LPAVDD808 pKa = 3.73YY809 pKa = 11.1HH810 pKa = 7.13PDD812 pKa = 3.4AEE814 pKa = 4.48ALVAAATGEE823 pKa = 4.14APDD826 pKa = 4.23RR827 pKa = 11.84NAVDD831 pKa = 3.96ALMTGSLAAEE841 pKa = 4.05WDD843 pKa = 3.72KK844 pKa = 10.91EE845 pKa = 4.33TLRR848 pKa = 11.84RR849 pKa = 11.84ILPRR853 pKa = 11.84SWIEE857 pKa = 3.56
Molecular weight: 94.6 kDa
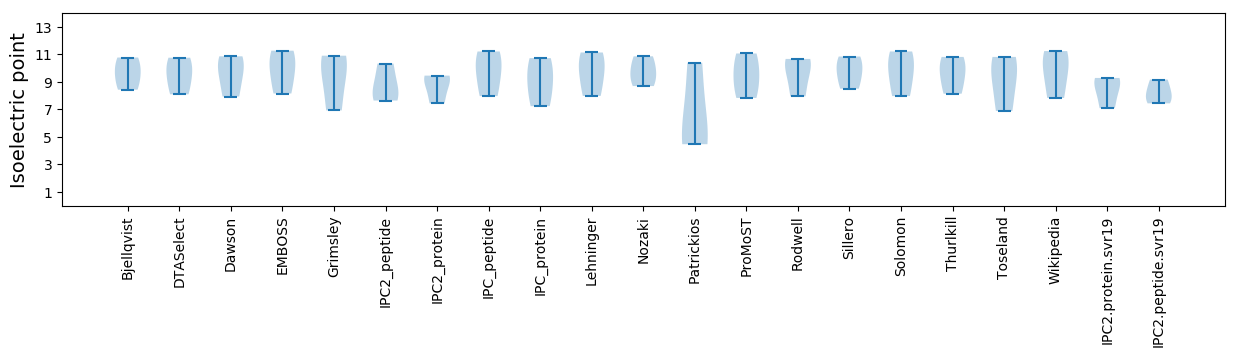
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLC4|A0A1L3KLC4_9VIRU Uncharacterized protein OS=Beihai weivirus-like virus 15 OX=1922743 PE=4 SV=1
MM1 pKa = 7.25WLWRR5 pKa = 11.84ACSRR9 pKa = 11.84DD10 pKa = 3.38SRR12 pKa = 11.84EE13 pKa = 3.75MEE15 pKa = 4.04ASVEE19 pKa = 4.13CARR22 pKa = 11.84FVTEE26 pKa = 4.67APLTRR31 pKa = 11.84WLTWMGISVWSTTCGPTASVVLWRR55 pKa = 11.84IGLRR59 pKa = 11.84SPAVIRR65 pKa = 11.84HH66 pKa = 4.67VARR69 pKa = 11.84LVGFCTLRR77 pKa = 11.84WKK79 pKa = 9.49MRR81 pKa = 11.84RR82 pKa = 11.84LSSTTTPVRR91 pKa = 11.84PMRR94 pKa = 11.84ALPPPLSTAYY104 pKa = 9.88GGSGAPRR111 pKa = 11.84SQLRR115 pKa = 11.84CCVCCLRR122 pKa = 11.84SLTQSRR128 pKa = 11.84LTVPIGMLCSTCGRR142 pKa = 11.84RR143 pKa = 11.84MAQAWSQRR151 pKa = 11.84NAHH154 pKa = 6.86LAAKK158 pKa = 9.49DD159 pKa = 3.67CVMEE163 pKa = 4.7LSSDD167 pKa = 3.77PPVGGFADD175 pKa = 3.84LPVWVNRR182 pKa = 11.84RR183 pKa = 11.84ILDD186 pKa = 3.4LRR188 pKa = 11.84GPTEE192 pKa = 3.92EE193 pKa = 5.38EE194 pKa = 3.52VWEE197 pKa = 4.16WVVQNSAPTRR207 pKa = 11.84RR208 pKa = 11.84PPMGRR213 pKa = 11.84RR214 pKa = 11.84TRR216 pKa = 11.84APPPPDD222 pKa = 3.91PYY224 pKa = 11.49GHH226 pKa = 7.38LEE228 pKa = 4.03GPVRR232 pKa = 11.84VAALIRR238 pKa = 11.84DD239 pKa = 4.01RR240 pKa = 11.84EE241 pKa = 4.1EE242 pKa = 3.23QAAARR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84QPGPLMRR256 pKa = 11.84LQEE259 pKa = 4.41KK260 pKa = 9.86EE261 pKa = 4.5RR262 pKa = 11.84EE263 pKa = 4.31AQSSSKK269 pKa = 11.11ADD271 pKa = 3.71LLARR275 pKa = 11.84GYY277 pKa = 11.29APLRR281 pKa = 11.84GPGAA285 pKa = 3.68
MM1 pKa = 7.25WLWRR5 pKa = 11.84ACSRR9 pKa = 11.84DD10 pKa = 3.38SRR12 pKa = 11.84EE13 pKa = 3.75MEE15 pKa = 4.04ASVEE19 pKa = 4.13CARR22 pKa = 11.84FVTEE26 pKa = 4.67APLTRR31 pKa = 11.84WLTWMGISVWSTTCGPTASVVLWRR55 pKa = 11.84IGLRR59 pKa = 11.84SPAVIRR65 pKa = 11.84HH66 pKa = 4.67VARR69 pKa = 11.84LVGFCTLRR77 pKa = 11.84WKK79 pKa = 9.49MRR81 pKa = 11.84RR82 pKa = 11.84LSSTTTPVRR91 pKa = 11.84PMRR94 pKa = 11.84ALPPPLSTAYY104 pKa = 9.88GGSGAPRR111 pKa = 11.84SQLRR115 pKa = 11.84CCVCCLRR122 pKa = 11.84SLTQSRR128 pKa = 11.84LTVPIGMLCSTCGRR142 pKa = 11.84RR143 pKa = 11.84MAQAWSQRR151 pKa = 11.84NAHH154 pKa = 6.86LAAKK158 pKa = 9.49DD159 pKa = 3.67CVMEE163 pKa = 4.7LSSDD167 pKa = 3.77PPVGGFADD175 pKa = 3.84LPVWVNRR182 pKa = 11.84RR183 pKa = 11.84ILDD186 pKa = 3.4LRR188 pKa = 11.84GPTEE192 pKa = 3.92EE193 pKa = 5.38EE194 pKa = 3.52VWEE197 pKa = 4.16WVVQNSAPTRR207 pKa = 11.84RR208 pKa = 11.84PPMGRR213 pKa = 11.84RR214 pKa = 11.84TRR216 pKa = 11.84APPPPDD222 pKa = 3.91PYY224 pKa = 11.49GHH226 pKa = 7.38LEE228 pKa = 4.03GPVRR232 pKa = 11.84VAALIRR238 pKa = 11.84DD239 pKa = 4.01RR240 pKa = 11.84EE241 pKa = 4.1EE242 pKa = 3.23QAAARR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84QPGPLMRR256 pKa = 11.84LQEE259 pKa = 4.41KK260 pKa = 9.86EE261 pKa = 4.5RR262 pKa = 11.84EE263 pKa = 4.31AQSSSKK269 pKa = 11.11ADD271 pKa = 3.71LLARR275 pKa = 11.84GYY277 pKa = 11.29APLRR281 pKa = 11.84GPGAA285 pKa = 3.68
Molecular weight: 31.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1497 |
285 |
857 |
499.0 |
54.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.957 ± 0.837 | 2.338 ± 0.656 |
5.144 ± 1.1 | 5.21 ± 0.748 |
2.004 ± 0.378 | 8.55 ± 0.948 |
1.937 ± 0.279 | 2.939 ± 0.436 |
3.273 ± 0.627 | 7.949 ± 0.722 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.071 ± 0.708 | 2.004 ± 0.378 |
7.749 ± 0.596 | 3.874 ± 0.494 |
9.686 ± 1.583 | 5.945 ± 0.693 |
6.346 ± 1.314 | 6.079 ± 0.393 |
2.338 ± 0.482 | 2.605 ± 0.524 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
