
Beihai weivirus-like virus 21
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.91
Get precalculated fractions of proteins
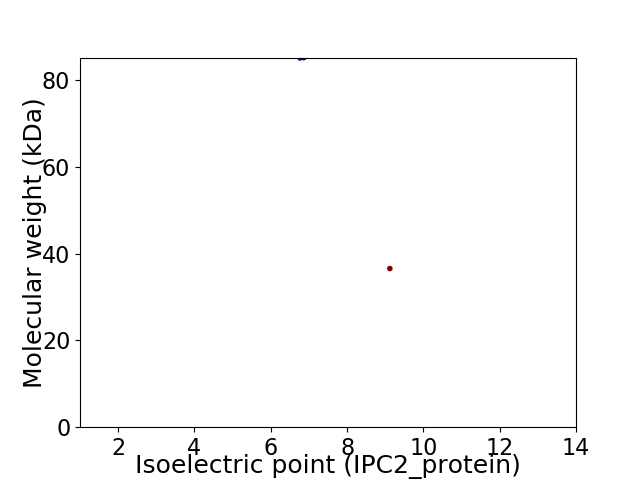
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
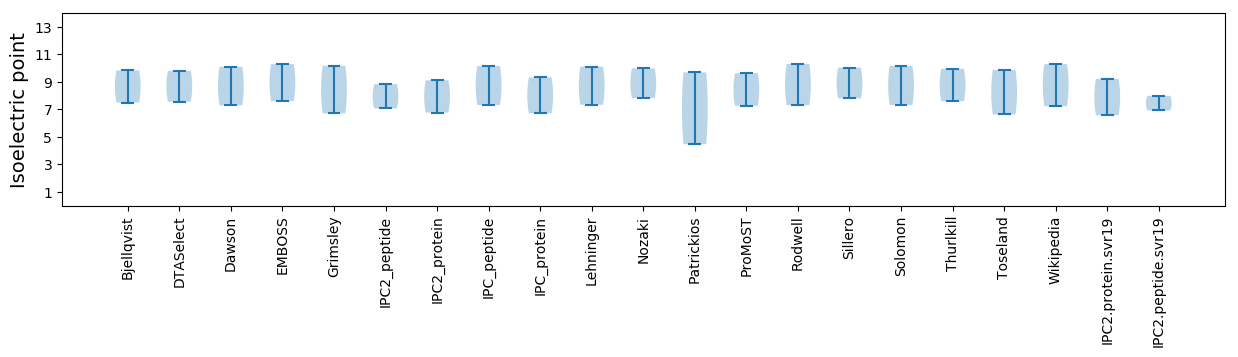
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL93|A0A1L3KL93_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 21 OX=1922750 PE=4 SV=1
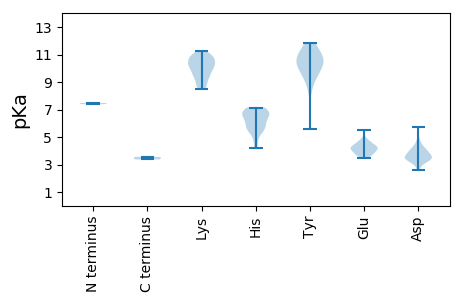
MM1 pKa = 7.39FGFGRR6 pKa = 11.84PALCSLCHH14 pKa = 6.6ADD16 pKa = 4.16EE17 pKa = 4.71LADD20 pKa = 3.76LSVPVGSDD28 pKa = 2.66NVYY31 pKa = 10.73RR32 pKa = 11.84KK33 pKa = 9.51MIIPANCSRR42 pKa = 11.84CRR44 pKa = 11.84HH45 pKa = 5.52SSVLSRR51 pKa = 11.84KK52 pKa = 9.28RR53 pKa = 11.84KK54 pKa = 8.65YY55 pKa = 9.48KK56 pKa = 10.28GHH58 pKa = 5.77MVYY61 pKa = 10.7LPARR65 pKa = 11.84LVADD69 pKa = 4.33ARR71 pKa = 11.84ALSAKK76 pKa = 10.28CGATTLWGMKK86 pKa = 8.79PANADD91 pKa = 3.57LQLVNLLSAEE101 pKa = 4.04RR102 pKa = 11.84DD103 pKa = 3.43GRR105 pKa = 11.84RR106 pKa = 11.84ANALLTVGLHH116 pKa = 6.63AIKK119 pKa = 10.7DD120 pKa = 3.81EE121 pKa = 4.06EE122 pKa = 4.58TFRR125 pKa = 11.84SGHH128 pKa = 4.68TCCVIDD134 pKa = 5.74ALIKK138 pKa = 10.83CLTDD142 pKa = 3.28QPRR145 pKa = 11.84NATSWRR151 pKa = 11.84RR152 pKa = 11.84VKK154 pKa = 8.79EE155 pKa = 4.56TPAKK159 pKa = 10.5AILDD163 pKa = 4.0GGKK166 pKa = 10.29GATDD170 pKa = 3.8DD171 pKa = 4.21ASVSGGAVSTGDD183 pKa = 3.96DD184 pKa = 3.51SSSIASAEE192 pKa = 3.83SAEE195 pKa = 4.26RR196 pKa = 11.84VNSQQSAFRR205 pKa = 11.84DD206 pKa = 3.6GTTIKK211 pKa = 9.6STVVEE216 pKa = 4.39APGTASTSGQAEE228 pKa = 4.28GEE230 pKa = 4.27EE231 pKa = 4.27VGARR235 pKa = 11.84VAQARR240 pKa = 11.84FPSMSEE246 pKa = 3.67KK247 pKa = 10.32EE248 pKa = 4.31VYY250 pKa = 10.45LFSNDD255 pKa = 3.1PRR257 pKa = 11.84NLEE260 pKa = 3.93AANTLRR266 pKa = 11.84NKK268 pKa = 10.3GVGVHH273 pKa = 5.48QPSEE277 pKa = 4.37KK278 pKa = 9.61EE279 pKa = 3.6AKK281 pKa = 10.38ARR283 pKa = 11.84DD284 pKa = 3.84EE285 pKa = 5.04IIAALCAQVFTEE297 pKa = 4.64RR298 pKa = 11.84EE299 pKa = 4.01CGKK302 pKa = 10.96AMMGYY307 pKa = 10.48EE308 pKa = 4.45SLTKK312 pKa = 9.68TALPRR317 pKa = 11.84KK318 pKa = 9.77LSEE321 pKa = 3.98QQKK324 pKa = 8.61EE325 pKa = 4.15QWQLDD330 pKa = 3.53AMKK333 pKa = 9.97EE334 pKa = 4.09AEE336 pKa = 4.51GDD338 pKa = 3.64GLSYY342 pKa = 11.33SKK344 pKa = 10.82FVDD347 pKa = 3.35AFVKK351 pKa = 10.9AEE353 pKa = 3.97VSAKK357 pKa = 10.0PKK359 pKa = 9.53PRR361 pKa = 11.84PIANHH366 pKa = 5.93KK367 pKa = 9.6EE368 pKa = 3.51IRR370 pKa = 11.84LCALAKK376 pKa = 9.5VAWVYY381 pKa = 10.89EE382 pKa = 3.9YY383 pKa = 11.83VLFHH387 pKa = 6.91KK388 pKa = 9.94FKK390 pKa = 10.94KK391 pKa = 10.0MSIKK395 pKa = 10.4HH396 pKa = 4.18RR397 pKa = 11.84TKK399 pKa = 10.6RR400 pKa = 11.84EE401 pKa = 3.95ALSDD405 pKa = 3.24IAGALSGMRR414 pKa = 11.84NGRR417 pKa = 11.84WCEE420 pKa = 3.56NDD422 pKa = 2.77LTAFEE427 pKa = 4.84FGISAKK433 pKa = 10.47LKK435 pKa = 9.13EE436 pKa = 4.61CEE438 pKa = 4.19CVILRR443 pKa = 11.84HH444 pKa = 5.67IASHH448 pKa = 6.54IGVEE452 pKa = 4.35DD453 pKa = 3.42VGPSLFEE460 pKa = 3.65RR461 pKa = 11.84VVNDD465 pKa = 3.42RR466 pKa = 11.84TKK468 pKa = 10.24ACVWSMRR475 pKa = 11.84YY476 pKa = 9.41KK477 pKa = 10.98DD478 pKa = 3.28EE479 pKa = 4.28TGAQRR484 pKa = 11.84KK485 pKa = 8.48FKK487 pKa = 11.2LEE489 pKa = 3.9LPNAMRR495 pKa = 11.84EE496 pKa = 4.17SGDD499 pKa = 3.52RR500 pKa = 11.84LTSSGNFLQNLIAWGSFLVEE520 pKa = 4.42PGHH523 pKa = 5.51VSKK526 pKa = 10.92AIEE529 pKa = 4.24SLVRR533 pKa = 11.84THH535 pKa = 6.7GEE537 pKa = 3.71KK538 pKa = 10.69LFYY541 pKa = 10.62TSARR545 pKa = 11.84DD546 pKa = 3.68GNKK549 pKa = 9.41YY550 pKa = 10.35LAMLVFEE557 pKa = 5.53GDD559 pKa = 3.46DD560 pKa = 3.26TLGRR564 pKa = 11.84LEE566 pKa = 4.11EE567 pKa = 4.54PVWEE571 pKa = 4.65AYY573 pKa = 10.11RR574 pKa = 11.84SGSEE578 pKa = 3.71TSIADD583 pKa = 3.76DD584 pKa = 4.63FFLRR588 pKa = 11.84WGWNPKK594 pKa = 10.11LSWKK598 pKa = 9.15ATTGHH603 pKa = 7.14DD604 pKa = 3.55YY605 pKa = 11.49ARR607 pKa = 11.84VVGYY611 pKa = 10.42DD612 pKa = 3.3VLIKK616 pKa = 10.92DD617 pKa = 4.11GVAVKK622 pKa = 10.85DD623 pKa = 3.38GDD625 pKa = 4.17SYY627 pKa = 10.45VACPEE632 pKa = 4.01MKK634 pKa = 10.32RR635 pKa = 11.84LLTTKK640 pKa = 9.52QWTTSSVTPEE650 pKa = 3.74EE651 pKa = 4.83LKK653 pKa = 8.89TCNRR657 pKa = 11.84IFAATLAADD666 pKa = 4.67FTRR669 pKa = 11.84VEE671 pKa = 3.99PFYY674 pKa = 11.51AFLKK678 pKa = 10.8SMYY681 pKa = 9.47EE682 pKa = 4.05SNRR685 pKa = 11.84GGKK688 pKa = 8.89NVSDD692 pKa = 3.39EE693 pKa = 4.25KK694 pKa = 11.24VRR696 pKa = 11.84EE697 pKa = 4.23FYY699 pKa = 11.02LATTGEE705 pKa = 4.27LPEE708 pKa = 5.34HH709 pKa = 6.62GSSKK713 pKa = 10.9LNDD716 pKa = 3.11IEE718 pKa = 4.65FPEE721 pKa = 4.36FNGTGSEE728 pKa = 3.87AWKK731 pKa = 10.0EE732 pKa = 3.79LARR735 pKa = 11.84VSCGDD740 pKa = 3.37FTDD743 pKa = 5.33AEE745 pKa = 4.36WATACAQPPHH755 pKa = 6.97DD756 pKa = 3.92RR757 pKa = 11.84HH758 pKa = 6.03GADD761 pKa = 4.14LAVGMPASWLAA772 pKa = 3.37
MM1 pKa = 7.39FGFGRR6 pKa = 11.84PALCSLCHH14 pKa = 6.6ADD16 pKa = 4.16EE17 pKa = 4.71LADD20 pKa = 3.76LSVPVGSDD28 pKa = 2.66NVYY31 pKa = 10.73RR32 pKa = 11.84KK33 pKa = 9.51MIIPANCSRR42 pKa = 11.84CRR44 pKa = 11.84HH45 pKa = 5.52SSVLSRR51 pKa = 11.84KK52 pKa = 9.28RR53 pKa = 11.84KK54 pKa = 8.65YY55 pKa = 9.48KK56 pKa = 10.28GHH58 pKa = 5.77MVYY61 pKa = 10.7LPARR65 pKa = 11.84LVADD69 pKa = 4.33ARR71 pKa = 11.84ALSAKK76 pKa = 10.28CGATTLWGMKK86 pKa = 8.79PANADD91 pKa = 3.57LQLVNLLSAEE101 pKa = 4.04RR102 pKa = 11.84DD103 pKa = 3.43GRR105 pKa = 11.84RR106 pKa = 11.84ANALLTVGLHH116 pKa = 6.63AIKK119 pKa = 10.7DD120 pKa = 3.81EE121 pKa = 4.06EE122 pKa = 4.58TFRR125 pKa = 11.84SGHH128 pKa = 4.68TCCVIDD134 pKa = 5.74ALIKK138 pKa = 10.83CLTDD142 pKa = 3.28QPRR145 pKa = 11.84NATSWRR151 pKa = 11.84RR152 pKa = 11.84VKK154 pKa = 8.79EE155 pKa = 4.56TPAKK159 pKa = 10.5AILDD163 pKa = 4.0GGKK166 pKa = 10.29GATDD170 pKa = 3.8DD171 pKa = 4.21ASVSGGAVSTGDD183 pKa = 3.96DD184 pKa = 3.51SSSIASAEE192 pKa = 3.83SAEE195 pKa = 4.26RR196 pKa = 11.84VNSQQSAFRR205 pKa = 11.84DD206 pKa = 3.6GTTIKK211 pKa = 9.6STVVEE216 pKa = 4.39APGTASTSGQAEE228 pKa = 4.28GEE230 pKa = 4.27EE231 pKa = 4.27VGARR235 pKa = 11.84VAQARR240 pKa = 11.84FPSMSEE246 pKa = 3.67KK247 pKa = 10.32EE248 pKa = 4.31VYY250 pKa = 10.45LFSNDD255 pKa = 3.1PRR257 pKa = 11.84NLEE260 pKa = 3.93AANTLRR266 pKa = 11.84NKK268 pKa = 10.3GVGVHH273 pKa = 5.48QPSEE277 pKa = 4.37KK278 pKa = 9.61EE279 pKa = 3.6AKK281 pKa = 10.38ARR283 pKa = 11.84DD284 pKa = 3.84EE285 pKa = 5.04IIAALCAQVFTEE297 pKa = 4.64RR298 pKa = 11.84EE299 pKa = 4.01CGKK302 pKa = 10.96AMMGYY307 pKa = 10.48EE308 pKa = 4.45SLTKK312 pKa = 9.68TALPRR317 pKa = 11.84KK318 pKa = 9.77LSEE321 pKa = 3.98QQKK324 pKa = 8.61EE325 pKa = 4.15QWQLDD330 pKa = 3.53AMKK333 pKa = 9.97EE334 pKa = 4.09AEE336 pKa = 4.51GDD338 pKa = 3.64GLSYY342 pKa = 11.33SKK344 pKa = 10.82FVDD347 pKa = 3.35AFVKK351 pKa = 10.9AEE353 pKa = 3.97VSAKK357 pKa = 10.0PKK359 pKa = 9.53PRR361 pKa = 11.84PIANHH366 pKa = 5.93KK367 pKa = 9.6EE368 pKa = 3.51IRR370 pKa = 11.84LCALAKK376 pKa = 9.5VAWVYY381 pKa = 10.89EE382 pKa = 3.9YY383 pKa = 11.83VLFHH387 pKa = 6.91KK388 pKa = 9.94FKK390 pKa = 10.94KK391 pKa = 10.0MSIKK395 pKa = 10.4HH396 pKa = 4.18RR397 pKa = 11.84TKK399 pKa = 10.6RR400 pKa = 11.84EE401 pKa = 3.95ALSDD405 pKa = 3.24IAGALSGMRR414 pKa = 11.84NGRR417 pKa = 11.84WCEE420 pKa = 3.56NDD422 pKa = 2.77LTAFEE427 pKa = 4.84FGISAKK433 pKa = 10.47LKK435 pKa = 9.13EE436 pKa = 4.61CEE438 pKa = 4.19CVILRR443 pKa = 11.84HH444 pKa = 5.67IASHH448 pKa = 6.54IGVEE452 pKa = 4.35DD453 pKa = 3.42VGPSLFEE460 pKa = 3.65RR461 pKa = 11.84VVNDD465 pKa = 3.42RR466 pKa = 11.84TKK468 pKa = 10.24ACVWSMRR475 pKa = 11.84YY476 pKa = 9.41KK477 pKa = 10.98DD478 pKa = 3.28EE479 pKa = 4.28TGAQRR484 pKa = 11.84KK485 pKa = 8.48FKK487 pKa = 11.2LEE489 pKa = 3.9LPNAMRR495 pKa = 11.84EE496 pKa = 4.17SGDD499 pKa = 3.52RR500 pKa = 11.84LTSSGNFLQNLIAWGSFLVEE520 pKa = 4.42PGHH523 pKa = 5.51VSKK526 pKa = 10.92AIEE529 pKa = 4.24SLVRR533 pKa = 11.84THH535 pKa = 6.7GEE537 pKa = 3.71KK538 pKa = 10.69LFYY541 pKa = 10.62TSARR545 pKa = 11.84DD546 pKa = 3.68GNKK549 pKa = 9.41YY550 pKa = 10.35LAMLVFEE557 pKa = 5.53GDD559 pKa = 3.46DD560 pKa = 3.26TLGRR564 pKa = 11.84LEE566 pKa = 4.11EE567 pKa = 4.54PVWEE571 pKa = 4.65AYY573 pKa = 10.11RR574 pKa = 11.84SGSEE578 pKa = 3.71TSIADD583 pKa = 3.76DD584 pKa = 4.63FFLRR588 pKa = 11.84WGWNPKK594 pKa = 10.11LSWKK598 pKa = 9.15ATTGHH603 pKa = 7.14DD604 pKa = 3.55YY605 pKa = 11.49ARR607 pKa = 11.84VVGYY611 pKa = 10.42DD612 pKa = 3.3VLIKK616 pKa = 10.92DD617 pKa = 4.11GVAVKK622 pKa = 10.85DD623 pKa = 3.38GDD625 pKa = 4.17SYY627 pKa = 10.45VACPEE632 pKa = 4.01MKK634 pKa = 10.32RR635 pKa = 11.84LLTTKK640 pKa = 9.52QWTTSSVTPEE650 pKa = 3.74EE651 pKa = 4.83LKK653 pKa = 8.89TCNRR657 pKa = 11.84IFAATLAADD666 pKa = 4.67FTRR669 pKa = 11.84VEE671 pKa = 3.99PFYY674 pKa = 11.51AFLKK678 pKa = 10.8SMYY681 pKa = 9.47EE682 pKa = 4.05SNRR685 pKa = 11.84GGKK688 pKa = 8.89NVSDD692 pKa = 3.39EE693 pKa = 4.25KK694 pKa = 11.24VRR696 pKa = 11.84EE697 pKa = 4.23FYY699 pKa = 11.02LATTGEE705 pKa = 4.27LPEE708 pKa = 5.34HH709 pKa = 6.62GSSKK713 pKa = 10.9LNDD716 pKa = 3.11IEE718 pKa = 4.65FPEE721 pKa = 4.36FNGTGSEE728 pKa = 3.87AWKK731 pKa = 10.0EE732 pKa = 3.79LARR735 pKa = 11.84VSCGDD740 pKa = 3.37FTDD743 pKa = 5.33AEE745 pKa = 4.36WATACAQPPHH755 pKa = 6.97DD756 pKa = 3.92RR757 pKa = 11.84HH758 pKa = 6.03GADD761 pKa = 4.14LAVGMPASWLAA772 pKa = 3.37
Molecular weight: 85.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL93|A0A1L3KL93_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 21 OX=1922750 PE=4 SV=1
MM1 pKa = 7.47ARR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84NTRR8 pKa = 11.84VTNGRR13 pKa = 11.84SGRR16 pKa = 11.84ARR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84PNAKK24 pKa = 9.34GASRR28 pKa = 11.84SVATRR33 pKa = 11.84VLATGVGASVPKK45 pKa = 10.76AFGSVAGYY53 pKa = 10.53GLHH56 pKa = 6.79CWDD59 pKa = 4.74AKK61 pKa = 10.73HH62 pKa = 6.29LAHH65 pKa = 6.98LPLPRR70 pKa = 11.84AVGPYY75 pKa = 5.61TTIRR79 pKa = 11.84ATRR82 pKa = 11.84RR83 pKa = 11.84IQISTHH89 pKa = 5.8ANVIGTFQCGSNMPEE104 pKa = 3.7QHH106 pKa = 6.53FDD108 pKa = 2.63WSEE111 pKa = 3.56AVMVTDD117 pKa = 4.1VVSGSAITAPSNAQLITLDD136 pKa = 4.14LGGLGDD142 pKa = 4.41AATLVPSALSVQIMCPTALQSASGIVYY169 pKa = 10.04AGVMNTQTAIAGRR182 pKa = 11.84NEE184 pKa = 3.81TWDD187 pKa = 3.25SYY189 pKa = 9.48MEE191 pKa = 4.73KK192 pKa = 10.08FVQFQNPRR200 pKa = 11.84LLAASKK206 pKa = 10.15LALRR210 pKa = 11.84GVQINSYY217 pKa = 8.46PLNMTEE223 pKa = 4.06VSKK226 pKa = 8.46FTPLCKK232 pKa = 10.18LADD235 pKa = 3.61SSAFTLDD242 pKa = 3.34GASIEE247 pKa = 4.24PAGWAPIVIFNPSGASLEE265 pKa = 4.19LLITMEE271 pKa = 3.65FRR273 pKa = 11.84VRR275 pKa = 11.84FDD277 pKa = 4.02LDD279 pKa = 3.43HH280 pKa = 7.02PASASHH286 pKa = 5.36THH288 pKa = 6.24HH289 pKa = 6.96PVASDD294 pKa = 3.75ALWDD298 pKa = 3.92RR299 pKa = 11.84MVKK302 pKa = 10.36GAVALGNGVVDD313 pKa = 3.66IADD316 pKa = 3.59VVANTGMAVGRR327 pKa = 11.84AVAVGSRR334 pKa = 11.84LAAASRR340 pKa = 11.84AMPALAVV347 pKa = 3.56
MM1 pKa = 7.47ARR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84NTRR8 pKa = 11.84VTNGRR13 pKa = 11.84SGRR16 pKa = 11.84ARR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84PNAKK24 pKa = 9.34GASRR28 pKa = 11.84SVATRR33 pKa = 11.84VLATGVGASVPKK45 pKa = 10.76AFGSVAGYY53 pKa = 10.53GLHH56 pKa = 6.79CWDD59 pKa = 4.74AKK61 pKa = 10.73HH62 pKa = 6.29LAHH65 pKa = 6.98LPLPRR70 pKa = 11.84AVGPYY75 pKa = 5.61TTIRR79 pKa = 11.84ATRR82 pKa = 11.84RR83 pKa = 11.84IQISTHH89 pKa = 5.8ANVIGTFQCGSNMPEE104 pKa = 3.7QHH106 pKa = 6.53FDD108 pKa = 2.63WSEE111 pKa = 3.56AVMVTDD117 pKa = 4.1VVSGSAITAPSNAQLITLDD136 pKa = 4.14LGGLGDD142 pKa = 4.41AATLVPSALSVQIMCPTALQSASGIVYY169 pKa = 10.04AGVMNTQTAIAGRR182 pKa = 11.84NEE184 pKa = 3.81TWDD187 pKa = 3.25SYY189 pKa = 9.48MEE191 pKa = 4.73KK192 pKa = 10.08FVQFQNPRR200 pKa = 11.84LLAASKK206 pKa = 10.15LALRR210 pKa = 11.84GVQINSYY217 pKa = 8.46PLNMTEE223 pKa = 4.06VSKK226 pKa = 8.46FTPLCKK232 pKa = 10.18LADD235 pKa = 3.61SSAFTLDD242 pKa = 3.34GASIEE247 pKa = 4.24PAGWAPIVIFNPSGASLEE265 pKa = 4.19LLITMEE271 pKa = 3.65FRR273 pKa = 11.84VRR275 pKa = 11.84FDD277 pKa = 4.02LDD279 pKa = 3.43HH280 pKa = 7.02PASASHH286 pKa = 5.36THH288 pKa = 6.24HH289 pKa = 6.96PVASDD294 pKa = 3.75ALWDD298 pKa = 3.92RR299 pKa = 11.84MVKK302 pKa = 10.36GAVALGNGVVDD313 pKa = 3.66IADD316 pKa = 3.59VVANTGMAVGRR327 pKa = 11.84AVAVGSRR334 pKa = 11.84LAAASRR340 pKa = 11.84AMPALAVV347 pKa = 3.56
Molecular weight: 36.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1119 |
347 |
772 |
559.5 |
60.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.332 ± 1.49 | 2.055 ± 0.507 |
5.094 ± 0.595 | 5.987 ± 2.068 |
3.485 ± 0.339 | 7.507 ± 0.316 |
2.324 ± 0.152 | 3.485 ± 0.47 |
5.451 ± 1.766 | 8.132 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.413 ± 0.425 | 3.485 ± 0.308 |
4.111 ± 0.604 | 2.324 ± 0.314 |
6.613 ± 0.17 | 8.132 ± 0.126 |
5.898 ± 0.41 | 7.328 ± 1.063 |
1.787 ± 0.195 | 2.055 ± 0.345 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
