
Hubei sobemo-like virus 29
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
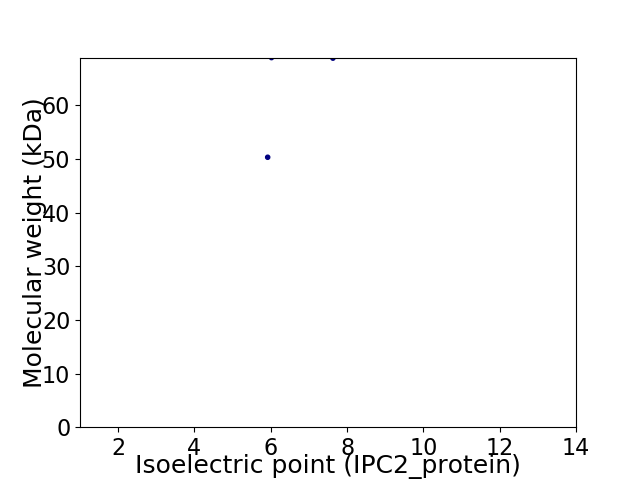
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEH6|A0A1L3KEH6_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 29 OX=1923215 PE=4 SV=1
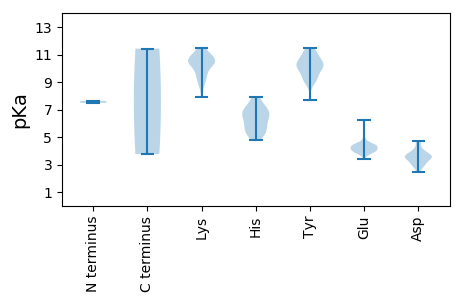
MM1 pKa = 7.63SGGEE5 pKa = 3.94PVRR8 pKa = 11.84FDD10 pKa = 3.86GFPEE14 pKa = 4.41EE15 pKa = 4.17YY16 pKa = 10.1RR17 pKa = 11.84EE18 pKa = 4.07QFEE21 pKa = 4.25KK22 pKa = 10.74AARR25 pKa = 11.84GYY27 pKa = 8.9GWPKK31 pKa = 9.88FGHH34 pKa = 5.69AAEE37 pKa = 4.48LRR39 pKa = 11.84SLHH42 pKa = 5.18YY43 pKa = 10.25HH44 pKa = 6.29ASLRR48 pKa = 11.84DD49 pKa = 3.55KK50 pKa = 10.37TRR52 pKa = 11.84SDD54 pKa = 3.64GPSRR58 pKa = 11.84EE59 pKa = 3.86ARR61 pKa = 11.84EE62 pKa = 4.46AIVQGVAEE70 pKa = 4.58QYY72 pKa = 11.4SSATWSIPEE81 pKa = 4.55DD82 pKa = 3.55FLQFSHH88 pKa = 7.12FEE90 pKa = 4.0RR91 pKa = 11.84VVKK94 pKa = 10.76NLDD97 pKa = 3.26WTSSPGYY104 pKa = 9.12PYY106 pKa = 9.87MRR108 pKa = 11.84RR109 pKa = 11.84APTNRR114 pKa = 11.84ILFNVNSIGVPDD126 pKa = 4.49PDD128 pKa = 3.34KK129 pKa = 11.29VSWMWEE135 pKa = 3.99VVQNQIRR142 pKa = 11.84DD143 pKa = 3.72RR144 pKa = 11.84VSDD147 pKa = 4.72PIRR150 pKa = 11.84LFIKK154 pKa = 10.71AEE156 pKa = 3.69PHH158 pKa = 5.74KK159 pKa = 10.7LKK161 pKa = 10.66KK162 pKa = 10.79LEE164 pKa = 4.4DD165 pKa = 3.18GRR167 pKa = 11.84YY168 pKa = 9.05RR169 pKa = 11.84LISSVSVVDD178 pKa = 3.47QLIDD182 pKa = 3.25HH183 pKa = 6.43MLFGDD188 pKa = 3.76MNEE191 pKa = 4.25SLVEE195 pKa = 3.74NWLNVPSKK203 pKa = 10.59IGWSHH208 pKa = 6.68LWGGWRR214 pKa = 11.84SIPRR218 pKa = 11.84DD219 pKa = 3.02WVAVDD224 pKa = 3.53KK225 pKa = 11.49SSWDD229 pKa = 2.93WTMQSWVCEE238 pKa = 3.92MVLEE242 pKa = 4.32VRR244 pKa = 11.84LRR246 pKa = 11.84LCSNPSQEE254 pKa = 4.44WLDD257 pKa = 3.68LAQWRR262 pKa = 11.84YY263 pKa = 10.18RR264 pKa = 11.84EE265 pKa = 4.5LFLNPLFVTSGGWILRR281 pKa = 11.84QMRR284 pKa = 11.84PGVMKK289 pKa = 10.25SGCVNTIADD298 pKa = 3.64NSIAQSILDD307 pKa = 3.37VRR309 pKa = 11.84VCYY312 pKa = 10.0EE313 pKa = 4.38LGLSPEE319 pKa = 4.47PLMSLGDD326 pKa = 3.42DD327 pKa = 4.13TYY329 pKa = 10.76QKK331 pKa = 10.6KK332 pKa = 9.02RR333 pKa = 11.84AAMGEE338 pKa = 4.02YY339 pKa = 10.41LEE341 pKa = 5.67RR342 pKa = 11.84LSQYY346 pKa = 11.16AILKK350 pKa = 9.12QVVEE354 pKa = 4.27APEE357 pKa = 3.91FGGFRR362 pKa = 11.84FLGPKK367 pKa = 9.01QVEE370 pKa = 3.76PLYY373 pKa = 10.25KK374 pKa = 10.37GKK376 pKa = 9.5HH377 pKa = 5.29AYY379 pKa = 9.17ILLHH383 pKa = 6.3VDD385 pKa = 3.49PEE387 pKa = 4.49VLPEE391 pKa = 4.29LAPSYY396 pKa = 10.31QLLYY400 pKa = 10.38HH401 pKa = 6.68RR402 pKa = 11.84SAYY405 pKa = 10.16RR406 pKa = 11.84DD407 pKa = 3.34MMEE410 pKa = 6.26DD411 pKa = 2.49IFLRR415 pKa = 11.84MGQEE419 pKa = 3.42IVPRR423 pKa = 11.84RR424 pKa = 11.84TRR426 pKa = 11.84DD427 pKa = 3.61LIFDD431 pKa = 4.62GYY433 pKa = 11.43
MM1 pKa = 7.63SGGEE5 pKa = 3.94PVRR8 pKa = 11.84FDD10 pKa = 3.86GFPEE14 pKa = 4.41EE15 pKa = 4.17YY16 pKa = 10.1RR17 pKa = 11.84EE18 pKa = 4.07QFEE21 pKa = 4.25KK22 pKa = 10.74AARR25 pKa = 11.84GYY27 pKa = 8.9GWPKK31 pKa = 9.88FGHH34 pKa = 5.69AAEE37 pKa = 4.48LRR39 pKa = 11.84SLHH42 pKa = 5.18YY43 pKa = 10.25HH44 pKa = 6.29ASLRR48 pKa = 11.84DD49 pKa = 3.55KK50 pKa = 10.37TRR52 pKa = 11.84SDD54 pKa = 3.64GPSRR58 pKa = 11.84EE59 pKa = 3.86ARR61 pKa = 11.84EE62 pKa = 4.46AIVQGVAEE70 pKa = 4.58QYY72 pKa = 11.4SSATWSIPEE81 pKa = 4.55DD82 pKa = 3.55FLQFSHH88 pKa = 7.12FEE90 pKa = 4.0RR91 pKa = 11.84VVKK94 pKa = 10.76NLDD97 pKa = 3.26WTSSPGYY104 pKa = 9.12PYY106 pKa = 9.87MRR108 pKa = 11.84RR109 pKa = 11.84APTNRR114 pKa = 11.84ILFNVNSIGVPDD126 pKa = 4.49PDD128 pKa = 3.34KK129 pKa = 11.29VSWMWEE135 pKa = 3.99VVQNQIRR142 pKa = 11.84DD143 pKa = 3.72RR144 pKa = 11.84VSDD147 pKa = 4.72PIRR150 pKa = 11.84LFIKK154 pKa = 10.71AEE156 pKa = 3.69PHH158 pKa = 5.74KK159 pKa = 10.7LKK161 pKa = 10.66KK162 pKa = 10.79LEE164 pKa = 4.4DD165 pKa = 3.18GRR167 pKa = 11.84YY168 pKa = 9.05RR169 pKa = 11.84LISSVSVVDD178 pKa = 3.47QLIDD182 pKa = 3.25HH183 pKa = 6.43MLFGDD188 pKa = 3.76MNEE191 pKa = 4.25SLVEE195 pKa = 3.74NWLNVPSKK203 pKa = 10.59IGWSHH208 pKa = 6.68LWGGWRR214 pKa = 11.84SIPRR218 pKa = 11.84DD219 pKa = 3.02WVAVDD224 pKa = 3.53KK225 pKa = 11.49SSWDD229 pKa = 2.93WTMQSWVCEE238 pKa = 3.92MVLEE242 pKa = 4.32VRR244 pKa = 11.84LRR246 pKa = 11.84LCSNPSQEE254 pKa = 4.44WLDD257 pKa = 3.68LAQWRR262 pKa = 11.84YY263 pKa = 10.18RR264 pKa = 11.84EE265 pKa = 4.5LFLNPLFVTSGGWILRR281 pKa = 11.84QMRR284 pKa = 11.84PGVMKK289 pKa = 10.25SGCVNTIADD298 pKa = 3.64NSIAQSILDD307 pKa = 3.37VRR309 pKa = 11.84VCYY312 pKa = 10.0EE313 pKa = 4.38LGLSPEE319 pKa = 4.47PLMSLGDD326 pKa = 3.42DD327 pKa = 4.13TYY329 pKa = 10.76QKK331 pKa = 10.6KK332 pKa = 9.02RR333 pKa = 11.84AAMGEE338 pKa = 4.02YY339 pKa = 10.41LEE341 pKa = 5.67RR342 pKa = 11.84LSQYY346 pKa = 11.16AILKK350 pKa = 9.12QVVEE354 pKa = 4.27APEE357 pKa = 3.91FGGFRR362 pKa = 11.84FLGPKK367 pKa = 9.01QVEE370 pKa = 3.76PLYY373 pKa = 10.25KK374 pKa = 10.37GKK376 pKa = 9.5HH377 pKa = 5.29AYY379 pKa = 9.17ILLHH383 pKa = 6.3VDD385 pKa = 3.49PEE387 pKa = 4.49VLPEE391 pKa = 4.29LAPSYY396 pKa = 10.31QLLYY400 pKa = 10.38HH401 pKa = 6.68RR402 pKa = 11.84SAYY405 pKa = 10.16RR406 pKa = 11.84DD407 pKa = 3.34MMEE410 pKa = 6.26DD411 pKa = 2.49IFLRR415 pKa = 11.84MGQEE419 pKa = 3.42IVPRR423 pKa = 11.84RR424 pKa = 11.84TRR426 pKa = 11.84DD427 pKa = 3.61LIFDD431 pKa = 4.62GYY433 pKa = 11.43
Molecular weight: 50.33 kDa
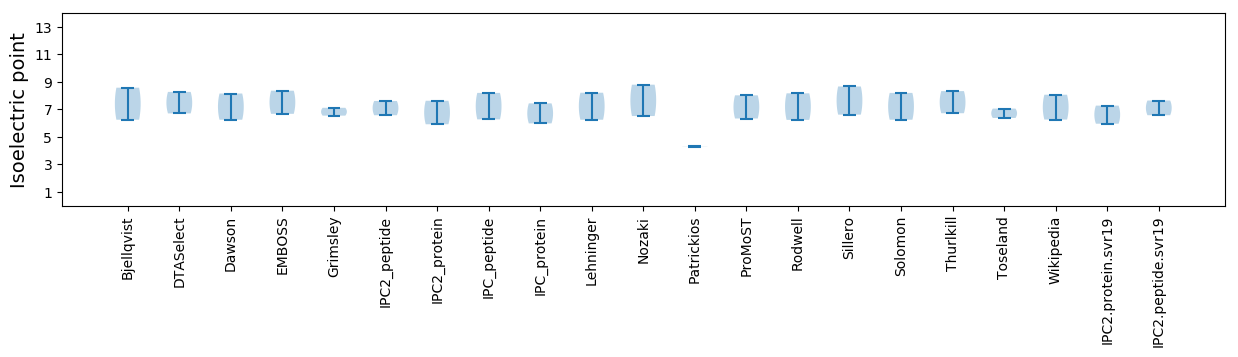
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEH6|A0A1L3KEH6_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 29 OX=1923215 PE=4 SV=1
MM1 pKa = 7.49GNRR4 pKa = 11.84VAVRR8 pKa = 11.84RR9 pKa = 11.84LEE11 pKa = 4.26DD12 pKa = 3.73EE13 pKa = 4.39EE14 pKa = 4.53TGSTGSGASPVYY26 pKa = 10.49AATTSPDD33 pKa = 3.01PNAWVNGVIWWCSKK47 pKa = 11.11ALLALLMVVTTAIVVYY63 pKa = 9.9RR64 pKa = 11.84WMAYY68 pKa = 9.39AVRR71 pKa = 11.84SATRR75 pKa = 11.84KK76 pKa = 9.38LRR78 pKa = 11.84SHH80 pKa = 5.9MEE82 pKa = 4.14EE83 pKa = 4.04IYY85 pKa = 11.15SLLDD89 pKa = 3.56YY90 pKa = 10.62ISPSEE95 pKa = 4.22TEE97 pKa = 3.88AEE99 pKa = 3.92RR100 pKa = 11.84EE101 pKa = 4.05EE102 pKa = 4.51KK103 pKa = 10.83VEE105 pKa = 3.81NVYY108 pKa = 10.45VRR110 pKa = 11.84VRR112 pKa = 11.84SAIEE116 pKa = 4.02DD117 pKa = 3.71LPIPSLLLTPSSDD130 pKa = 2.82WVFLAFGVVTLLVGCYY146 pKa = 9.44VSMRR150 pKa = 11.84SFGRR154 pKa = 11.84VSRR157 pKa = 11.84NLVHH161 pKa = 7.93RR162 pKa = 11.84IRR164 pKa = 11.84GIQYY168 pKa = 7.68EE169 pKa = 4.36SVRR172 pKa = 11.84EE173 pKa = 4.05GSSFRR178 pKa = 11.84SAKK181 pKa = 9.82VPAYY185 pKa = 10.1QIAVMEE191 pKa = 5.0AGLLSDD197 pKa = 2.89THH199 pKa = 7.77IGYY202 pKa = 9.6GCRR205 pKa = 11.84YY206 pKa = 9.29CDD208 pKa = 3.64YY209 pKa = 10.81LVLPRR214 pKa = 11.84HH215 pKa = 5.44VLEE218 pKa = 4.82KK219 pKa = 10.93DD220 pKa = 3.11GALVTTVLLKK230 pKa = 11.07GPLSKK235 pKa = 11.17VMTNISPVQSRR246 pKa = 11.84VIDD249 pKa = 3.98DD250 pKa = 4.73LVYY253 pKa = 10.72CYY255 pKa = 10.6LEE257 pKa = 4.34PKK259 pKa = 8.63TWSCLGAAKK268 pKa = 10.27GKK270 pKa = 8.66WANKK274 pKa = 9.5LMSLHH279 pKa = 5.25VTCTGIQGQTAGRR292 pKa = 11.84LARR295 pKa = 11.84TAIRR299 pKa = 11.84WMMSYY304 pKa = 9.67TGTTIEE310 pKa = 4.31GMSGAAYY317 pKa = 9.02EE318 pKa = 4.5SQGEE322 pKa = 3.91IHH324 pKa = 7.28GIHH327 pKa = 6.06QGATGPFNLGISSLLVTQEE346 pKa = 3.84MALLCQVEE354 pKa = 4.46SSLDD358 pKa = 3.53TNKK361 pKa = 10.4DD362 pKa = 2.89AKK364 pKa = 11.16VPLFYY369 pKa = 11.09NQANAKK375 pKa = 9.26LWSQIEE381 pKa = 4.12AMDD384 pKa = 4.17EE385 pKa = 3.78LLEE388 pKa = 4.69RR389 pKa = 11.84YY390 pKa = 10.09SSDD393 pKa = 2.45SWALGGEE400 pKa = 3.93VDD402 pKa = 4.05YY403 pKa = 11.45SKK405 pKa = 11.48KK406 pKa = 10.64LDD408 pKa = 3.93FGEE411 pKa = 4.07EE412 pKa = 3.59AARR415 pKa = 11.84RR416 pKa = 11.84RR417 pKa = 11.84PVSKK421 pKa = 10.33SHH423 pKa = 7.02LNVTLPEE430 pKa = 4.31GGIRR434 pKa = 11.84LANQNNTGSTTDD446 pKa = 3.62YY447 pKa = 11.22VVVTSAEE454 pKa = 4.21KK455 pKa = 10.48EE456 pKa = 3.97FLMEE460 pKa = 4.93LGRR463 pKa = 11.84SAILEE468 pKa = 4.31RR469 pKa = 11.84IEE471 pKa = 4.35ALEE474 pKa = 4.24TKK476 pKa = 10.14VAALGKK482 pKa = 10.48AEE484 pKa = 4.47IPCPHH489 pKa = 7.15CEE491 pKa = 4.13TTCRR495 pKa = 11.84TRR497 pKa = 11.84EE498 pKa = 4.01RR499 pKa = 11.84MDD501 pKa = 2.94HH502 pKa = 6.66HH503 pKa = 6.87IASAHH508 pKa = 5.37PKK510 pKa = 9.45VYY512 pKa = 9.94RR513 pKa = 11.84CQFCEE518 pKa = 4.32VSCRR522 pKa = 11.84TEE524 pKa = 3.97KK525 pKa = 10.86KK526 pKa = 10.21LQNHH530 pKa = 6.72LEE532 pKa = 4.14NNHH535 pKa = 4.76VVKK538 pKa = 10.76KK539 pKa = 10.1EE540 pKa = 3.87SAIPADD546 pKa = 3.62TGKK549 pKa = 10.33SGKK552 pKa = 7.93VVKK555 pKa = 9.74TGSFLGKK562 pKa = 9.9RR563 pKa = 11.84QGSQRR568 pKa = 11.84NNTVSSARR576 pKa = 11.84SSSLSSRR583 pKa = 11.84APPSPCLEE591 pKa = 4.08EE592 pKa = 4.36SLSGLMASQRR602 pKa = 11.84SIEE605 pKa = 4.13SSLKK609 pKa = 10.45KK610 pKa = 10.34LLEE613 pKa = 4.11VMAGRR618 pKa = 11.84NSAMQQSS625 pKa = 3.76
MM1 pKa = 7.49GNRR4 pKa = 11.84VAVRR8 pKa = 11.84RR9 pKa = 11.84LEE11 pKa = 4.26DD12 pKa = 3.73EE13 pKa = 4.39EE14 pKa = 4.53TGSTGSGASPVYY26 pKa = 10.49AATTSPDD33 pKa = 3.01PNAWVNGVIWWCSKK47 pKa = 11.11ALLALLMVVTTAIVVYY63 pKa = 9.9RR64 pKa = 11.84WMAYY68 pKa = 9.39AVRR71 pKa = 11.84SATRR75 pKa = 11.84KK76 pKa = 9.38LRR78 pKa = 11.84SHH80 pKa = 5.9MEE82 pKa = 4.14EE83 pKa = 4.04IYY85 pKa = 11.15SLLDD89 pKa = 3.56YY90 pKa = 10.62ISPSEE95 pKa = 4.22TEE97 pKa = 3.88AEE99 pKa = 3.92RR100 pKa = 11.84EE101 pKa = 4.05EE102 pKa = 4.51KK103 pKa = 10.83VEE105 pKa = 3.81NVYY108 pKa = 10.45VRR110 pKa = 11.84VRR112 pKa = 11.84SAIEE116 pKa = 4.02DD117 pKa = 3.71LPIPSLLLTPSSDD130 pKa = 2.82WVFLAFGVVTLLVGCYY146 pKa = 9.44VSMRR150 pKa = 11.84SFGRR154 pKa = 11.84VSRR157 pKa = 11.84NLVHH161 pKa = 7.93RR162 pKa = 11.84IRR164 pKa = 11.84GIQYY168 pKa = 7.68EE169 pKa = 4.36SVRR172 pKa = 11.84EE173 pKa = 4.05GSSFRR178 pKa = 11.84SAKK181 pKa = 9.82VPAYY185 pKa = 10.1QIAVMEE191 pKa = 5.0AGLLSDD197 pKa = 2.89THH199 pKa = 7.77IGYY202 pKa = 9.6GCRR205 pKa = 11.84YY206 pKa = 9.29CDD208 pKa = 3.64YY209 pKa = 10.81LVLPRR214 pKa = 11.84HH215 pKa = 5.44VLEE218 pKa = 4.82KK219 pKa = 10.93DD220 pKa = 3.11GALVTTVLLKK230 pKa = 11.07GPLSKK235 pKa = 11.17VMTNISPVQSRR246 pKa = 11.84VIDD249 pKa = 3.98DD250 pKa = 4.73LVYY253 pKa = 10.72CYY255 pKa = 10.6LEE257 pKa = 4.34PKK259 pKa = 8.63TWSCLGAAKK268 pKa = 10.27GKK270 pKa = 8.66WANKK274 pKa = 9.5LMSLHH279 pKa = 5.25VTCTGIQGQTAGRR292 pKa = 11.84LARR295 pKa = 11.84TAIRR299 pKa = 11.84WMMSYY304 pKa = 9.67TGTTIEE310 pKa = 4.31GMSGAAYY317 pKa = 9.02EE318 pKa = 4.5SQGEE322 pKa = 3.91IHH324 pKa = 7.28GIHH327 pKa = 6.06QGATGPFNLGISSLLVTQEE346 pKa = 3.84MALLCQVEE354 pKa = 4.46SSLDD358 pKa = 3.53TNKK361 pKa = 10.4DD362 pKa = 2.89AKK364 pKa = 11.16VPLFYY369 pKa = 11.09NQANAKK375 pKa = 9.26LWSQIEE381 pKa = 4.12AMDD384 pKa = 4.17EE385 pKa = 3.78LLEE388 pKa = 4.69RR389 pKa = 11.84YY390 pKa = 10.09SSDD393 pKa = 2.45SWALGGEE400 pKa = 3.93VDD402 pKa = 4.05YY403 pKa = 11.45SKK405 pKa = 11.48KK406 pKa = 10.64LDD408 pKa = 3.93FGEE411 pKa = 4.07EE412 pKa = 3.59AARR415 pKa = 11.84RR416 pKa = 11.84RR417 pKa = 11.84PVSKK421 pKa = 10.33SHH423 pKa = 7.02LNVTLPEE430 pKa = 4.31GGIRR434 pKa = 11.84LANQNNTGSTTDD446 pKa = 3.62YY447 pKa = 11.22VVVTSAEE454 pKa = 4.21KK455 pKa = 10.48EE456 pKa = 3.97FLMEE460 pKa = 4.93LGRR463 pKa = 11.84SAILEE468 pKa = 4.31RR469 pKa = 11.84IEE471 pKa = 4.35ALEE474 pKa = 4.24TKK476 pKa = 10.14VAALGKK482 pKa = 10.48AEE484 pKa = 4.47IPCPHH489 pKa = 7.15CEE491 pKa = 4.13TTCRR495 pKa = 11.84TRR497 pKa = 11.84EE498 pKa = 4.01RR499 pKa = 11.84MDD501 pKa = 2.94HH502 pKa = 6.66HH503 pKa = 6.87IASAHH508 pKa = 5.37PKK510 pKa = 9.45VYY512 pKa = 9.94RR513 pKa = 11.84CQFCEE518 pKa = 4.32VSCRR522 pKa = 11.84TEE524 pKa = 3.97KK525 pKa = 10.86KK526 pKa = 10.21LQNHH530 pKa = 6.72LEE532 pKa = 4.14NNHH535 pKa = 4.76VVKK538 pKa = 10.76KK539 pKa = 10.1EE540 pKa = 3.87SAIPADD546 pKa = 3.62TGKK549 pKa = 10.33SGKK552 pKa = 7.93VVKK555 pKa = 9.74TGSFLGKK562 pKa = 9.9RR563 pKa = 11.84QGSQRR568 pKa = 11.84NNTVSSARR576 pKa = 11.84SSSLSSRR583 pKa = 11.84APPSPCLEE591 pKa = 4.08EE592 pKa = 4.36SLSGLMASQRR602 pKa = 11.84SIEE605 pKa = 4.13SSLKK609 pKa = 10.45KK610 pKa = 10.34LLEE613 pKa = 4.11VMAGRR618 pKa = 11.84NSAMQQSS625 pKa = 3.76
Molecular weight: 68.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1058 |
433 |
625 |
529.0 |
59.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.994 ± 1.164 | 1.796 ± 0.53 |
4.253 ± 1.065 | 7.278 ± 0.072 |
2.552 ± 0.836 | 6.711 ± 0.149 |
2.268 ± 0.025 | 4.348 ± 0.165 |
4.631 ± 0.288 | 9.735 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.025 ± 0.127 | 3.119 ± 0.211 |
4.631 ± 0.835 | 3.497 ± 0.401 |
7.183 ± 0.547 | 9.546 ± 0.89 |
4.442 ± 1.438 | 7.845 ± 0.277 |
2.457 ± 0.753 | 3.686 ± 0.286 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
