
Fistulifera solaris (Oleaginous diatom)
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Ochrophyta; Bacillariophyta; Bacillariophyceae; Bacillariophycidae; Naviculales; Naviculaceae; Fistulifera
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
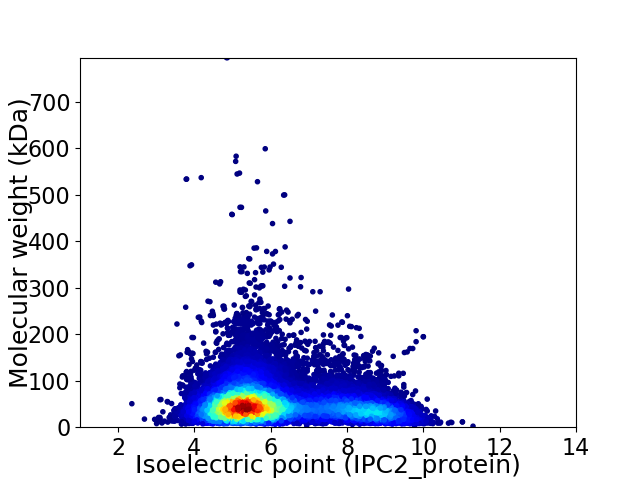
Virtual 2D-PAGE plot for 20319 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z5KJQ0|A0A1Z5KJQ0_FISSO Uncharacterized protein OS=Fistulifera solaris OX=1519565 GN=FisN_16Lh186 PE=4 SV=1
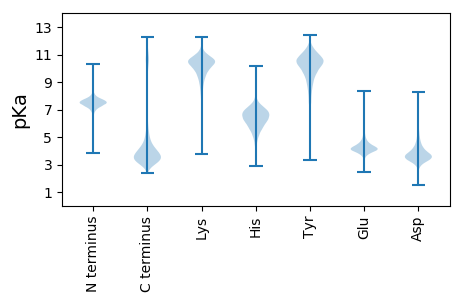
MM1 pKa = 7.85KK2 pKa = 9.1ITTFATSLLLASVLVPNQASQVDD25 pKa = 3.71EE26 pKa = 4.48ALPHH30 pKa = 5.48FRR32 pKa = 11.84ARR34 pKa = 11.84SLLTAVEE41 pKa = 4.5LGNGAQLSVNINAPATGSSVVVDD64 pKa = 4.17DD65 pKa = 4.19TCAASTPFDD74 pKa = 3.45GSASVGKK81 pKa = 10.36GNPDD85 pKa = 2.69VTYY88 pKa = 10.65IFIIDD93 pKa = 3.81EE94 pKa = 4.45SGSTSGIIAGIKK106 pKa = 10.44AFFEE110 pKa = 4.63DD111 pKa = 3.25LTDD114 pKa = 3.41IVFSEE119 pKa = 5.16GSALNAGVVRR129 pKa = 11.84FGSSAIRR136 pKa = 11.84MSALTDD142 pKa = 4.51DD143 pKa = 3.39IAAIKK148 pKa = 10.4DD149 pKa = 3.83VIDD152 pKa = 3.64NPFLNGPSTNCADD165 pKa = 3.97SLVKK169 pKa = 10.52ARR171 pKa = 11.84EE172 pKa = 3.88LAEE175 pKa = 4.06TSTAATTIVLFAGDD189 pKa = 4.5GACNSGGSFVGPANDD204 pKa = 4.24LAGLANTNVIVEE216 pKa = 4.77TIAIGIGCQGDD227 pKa = 3.91LGSIPRR233 pKa = 11.84NGGSCKK239 pKa = 10.42AVTSVNDD246 pKa = 3.3FDD248 pKa = 3.59ITTVIGTTLKK258 pKa = 10.85DD259 pKa = 3.12VAYY262 pKa = 9.85NLNAGAFASLTTSPSGDD279 pKa = 3.29TAGPATKK286 pKa = 9.75TFVQAIPINLGPNDD300 pKa = 4.37LCVKK304 pKa = 9.77ATGNDD309 pKa = 3.09EE310 pKa = 4.78VGQQDD315 pKa = 3.65VDD317 pKa = 3.81VLEE320 pKa = 5.36CISIVGVDD328 pKa = 3.85ASAPTITCPANIITATDD345 pKa = 3.21AGLCEE350 pKa = 4.48SSVATGSATAIDD362 pKa = 3.98NCSDD366 pKa = 3.47GLSPQASATGPFPLGISTVTFSVVDD391 pKa = 3.65ASGNPASCDD400 pKa = 3.33MSVEE404 pKa = 4.29VYY406 pKa = 10.62DD407 pKa = 3.95GEE409 pKa = 4.53EE410 pKa = 4.52PILLCPADD418 pKa = 3.72MTVSTDD424 pKa = 3.86AGICTATIEE433 pKa = 4.38IPVATVTDD441 pKa = 3.66NCDD444 pKa = 3.07QGLTPSTTAVNPFALGSTTVEE465 pKa = 3.92YY466 pKa = 9.95TVADD470 pKa = 3.6NAGNTDD476 pKa = 2.94ACTFAVVVEE485 pKa = 4.61DD486 pKa = 4.39NEE488 pKa = 4.63APTATCSPANNPGGNVPKK506 pKa = 10.72ANNQDD511 pKa = 3.27GFFVLGGEE519 pKa = 4.62DD520 pKa = 3.36NCSVLSYY527 pKa = 10.58QVVDD531 pKa = 3.93SVSGFVFGPFPSGTVFKK548 pKa = 9.86YY549 pKa = 9.47TEE551 pKa = 4.58APGSTPSQKK560 pKa = 10.39PGTGNVNYY568 pKa = 9.75FLKK571 pKa = 10.89GKK573 pKa = 10.03GDD575 pKa = 3.76AYY577 pKa = 11.63VLVTDD582 pKa = 4.9GAGNTAMAPCLVPPKK597 pKa = 9.86PSRR600 pKa = 11.84LLRR603 pKa = 11.84GII605 pKa = 4.15
MM1 pKa = 7.85KK2 pKa = 9.1ITTFATSLLLASVLVPNQASQVDD25 pKa = 3.71EE26 pKa = 4.48ALPHH30 pKa = 5.48FRR32 pKa = 11.84ARR34 pKa = 11.84SLLTAVEE41 pKa = 4.5LGNGAQLSVNINAPATGSSVVVDD64 pKa = 4.17DD65 pKa = 4.19TCAASTPFDD74 pKa = 3.45GSASVGKK81 pKa = 10.36GNPDD85 pKa = 2.69VTYY88 pKa = 10.65IFIIDD93 pKa = 3.81EE94 pKa = 4.45SGSTSGIIAGIKK106 pKa = 10.44AFFEE110 pKa = 4.63DD111 pKa = 3.25LTDD114 pKa = 3.41IVFSEE119 pKa = 5.16GSALNAGVVRR129 pKa = 11.84FGSSAIRR136 pKa = 11.84MSALTDD142 pKa = 4.51DD143 pKa = 3.39IAAIKK148 pKa = 10.4DD149 pKa = 3.83VIDD152 pKa = 3.64NPFLNGPSTNCADD165 pKa = 3.97SLVKK169 pKa = 10.52ARR171 pKa = 11.84EE172 pKa = 3.88LAEE175 pKa = 4.06TSTAATTIVLFAGDD189 pKa = 4.5GACNSGGSFVGPANDD204 pKa = 4.24LAGLANTNVIVEE216 pKa = 4.77TIAIGIGCQGDD227 pKa = 3.91LGSIPRR233 pKa = 11.84NGGSCKK239 pKa = 10.42AVTSVNDD246 pKa = 3.3FDD248 pKa = 3.59ITTVIGTTLKK258 pKa = 10.85DD259 pKa = 3.12VAYY262 pKa = 9.85NLNAGAFASLTTSPSGDD279 pKa = 3.29TAGPATKK286 pKa = 9.75TFVQAIPINLGPNDD300 pKa = 4.37LCVKK304 pKa = 9.77ATGNDD309 pKa = 3.09EE310 pKa = 4.78VGQQDD315 pKa = 3.65VDD317 pKa = 3.81VLEE320 pKa = 5.36CISIVGVDD328 pKa = 3.85ASAPTITCPANIITATDD345 pKa = 3.21AGLCEE350 pKa = 4.48SSVATGSATAIDD362 pKa = 3.98NCSDD366 pKa = 3.47GLSPQASATGPFPLGISTVTFSVVDD391 pKa = 3.65ASGNPASCDD400 pKa = 3.33MSVEE404 pKa = 4.29VYY406 pKa = 10.62DD407 pKa = 3.95GEE409 pKa = 4.53EE410 pKa = 4.52PILLCPADD418 pKa = 3.72MTVSTDD424 pKa = 3.86AGICTATIEE433 pKa = 4.38IPVATVTDD441 pKa = 3.66NCDD444 pKa = 3.07QGLTPSTTAVNPFALGSTTVEE465 pKa = 3.92YY466 pKa = 9.95TVADD470 pKa = 3.6NAGNTDD476 pKa = 2.94ACTFAVVVEE485 pKa = 4.61DD486 pKa = 4.39NEE488 pKa = 4.63APTATCSPANNPGGNVPKK506 pKa = 10.72ANNQDD511 pKa = 3.27GFFVLGGEE519 pKa = 4.62DD520 pKa = 3.36NCSVLSYY527 pKa = 10.58QVVDD531 pKa = 3.93SVSGFVFGPFPSGTVFKK548 pKa = 9.86YY549 pKa = 9.47TEE551 pKa = 4.58APGSTPSQKK560 pKa = 10.39PGTGNVNYY568 pKa = 9.75FLKK571 pKa = 10.89GKK573 pKa = 10.03GDD575 pKa = 3.76AYY577 pKa = 11.63VLVTDD582 pKa = 4.9GAGNTAMAPCLVPPKK597 pKa = 9.86PSRR600 pKa = 11.84LLRR603 pKa = 11.84GII605 pKa = 4.15
Molecular weight: 60.97 kDa
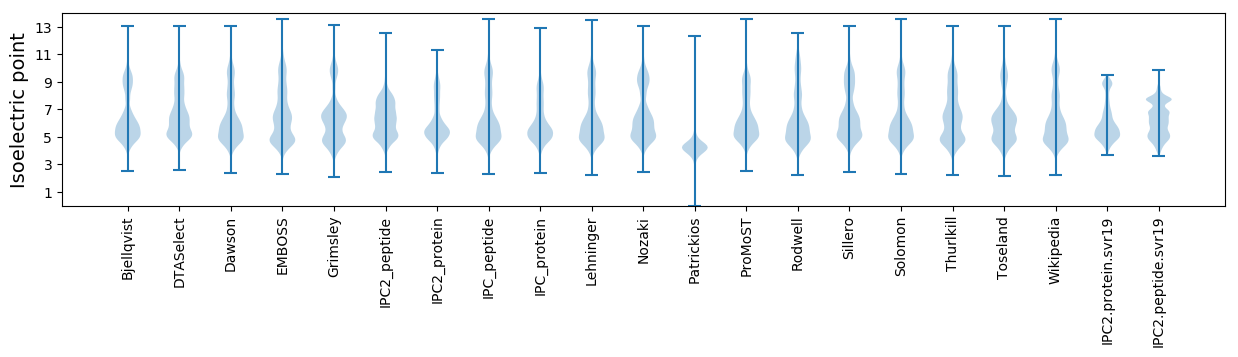
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z5KD27|A0A1Z5KD27_FISSO Isoform of A0A1Z5KCW6 Uncharacterized protein (Fragment) OS=Fistulifera solaris OX=1519565 GN=FisN_26Lh116 PE=4 SV=1
RR1 pKa = 7.04RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SS14 pKa = 3.02
RR1 pKa = 7.04RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SS14 pKa = 3.02
Molecular weight: 2.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
10061793 |
14 |
7171 |
495.2 |
55.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.031 ± 0.014 | 1.622 ± 0.008 |
6.036 ± 0.013 | 6.699 ± 0.018 |
3.845 ± 0.011 | 5.942 ± 0.015 |
2.406 ± 0.009 | 4.721 ± 0.008 |
5.108 ± 0.016 | 9.451 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.404 ± 0.006 | 3.894 ± 0.008 |
5.081 ± 0.017 | 4.423 ± 0.014 |
5.764 ± 0.012 | 8.275 ± 0.021 |
5.83 ± 0.012 | 6.493 ± 0.011 |
1.318 ± 0.006 | 2.657 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
