
Medicago sativa amalgavirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; unclassified Amalgaviridae
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
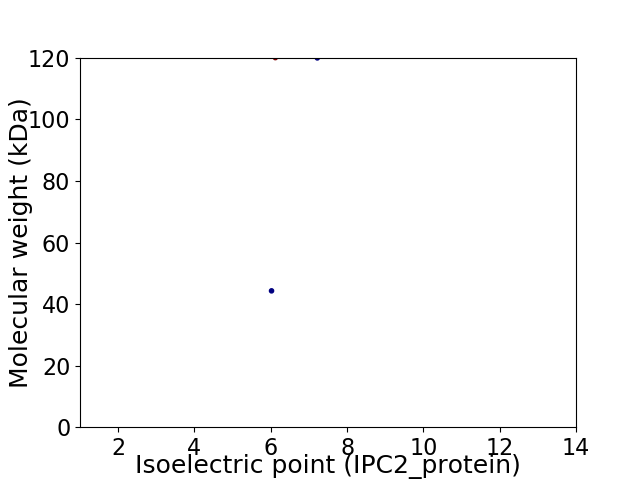
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
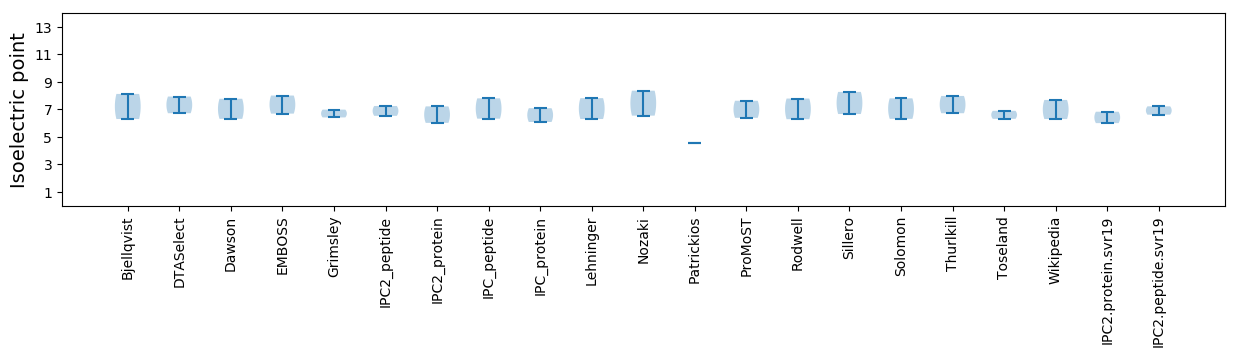
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V0A0B8|A0A2V0A0B8_9VIRU ORF1+2p OS=Medicago sativa amalgavirus 1 OX=2069326 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.95ADD3 pKa = 4.38FEE5 pKa = 4.39AQRR8 pKa = 11.84DD9 pKa = 3.92DD10 pKa = 5.06NIAANAPIGGLSSKK24 pKa = 8.01QQEE27 pKa = 4.29IDD29 pKa = 3.4TVTAAITPLLAAGFPQAIFNYY50 pKa = 10.32DD51 pKa = 3.5DD52 pKa = 4.24LLLRR56 pKa = 11.84GYY58 pKa = 7.12TAKK61 pKa = 10.03TFCDD65 pKa = 5.23FIKK68 pKa = 10.16PLSAITEE75 pKa = 4.04RR76 pKa = 11.84RR77 pKa = 11.84EE78 pKa = 4.35LISLCALGNNRR89 pKa = 11.84FWDD92 pKa = 3.6MSVVAEE98 pKa = 4.57LDD100 pKa = 3.35EE101 pKa = 4.34FLNFVKK107 pKa = 10.02WLKK110 pKa = 10.79SPEE113 pKa = 3.9GRR115 pKa = 11.84DD116 pKa = 3.51AQTQAAKK123 pKa = 10.32KK124 pKa = 9.67RR125 pKa = 11.84ALNKK129 pKa = 9.82KK130 pKa = 9.92ASDD133 pKa = 3.5GMSTKK138 pKa = 10.57DD139 pKa = 3.32VALVQMGNAIIADD152 pKa = 3.74YY153 pKa = 10.58QRR155 pKa = 11.84EE156 pKa = 4.18RR157 pKa = 11.84KK158 pKa = 8.4QRR160 pKa = 11.84RR161 pKa = 11.84FPIEE165 pKa = 3.64EE166 pKa = 3.69EE167 pKa = 3.84MAEE170 pKa = 4.25LRR172 pKa = 11.84RR173 pKa = 11.84QLRR176 pKa = 11.84QLDD179 pKa = 4.0EE180 pKa = 4.07EE181 pKa = 4.34LQAVEE186 pKa = 4.6EE187 pKa = 4.45EE188 pKa = 4.43IKK190 pKa = 11.01VKK192 pKa = 10.49YY193 pKa = 10.57GPVALYY199 pKa = 8.86EE200 pKa = 4.64GPDD203 pKa = 3.4NTRR206 pKa = 11.84VKK208 pKa = 10.78SDD210 pKa = 3.59AYY212 pKa = 11.43LMYY215 pKa = 10.52QDD217 pKa = 4.71DD218 pKa = 4.42CRR220 pKa = 11.84KK221 pKa = 9.74KK222 pKa = 9.78GYY224 pKa = 9.7RR225 pKa = 11.84AIAQYY230 pKa = 10.71QGGFEE235 pKa = 4.14KK236 pKa = 10.62AVEE239 pKa = 3.94LFGNKK244 pKa = 7.29VRR246 pKa = 11.84EE247 pKa = 4.21AHH249 pKa = 5.71FCAYY253 pKa = 10.48LSDD256 pKa = 4.01PARR259 pKa = 11.84SDD261 pKa = 3.67FVRR264 pKa = 11.84DD265 pKa = 3.53YY266 pKa = 9.79YY267 pKa = 10.07NQKK270 pKa = 9.46IVHH273 pKa = 6.41LEE275 pKa = 3.55RR276 pKa = 11.84SGEE279 pKa = 4.08KK280 pKa = 9.92KK281 pKa = 9.73QAGSFRR287 pKa = 11.84SLLVAAHH294 pKa = 6.63GEE296 pKa = 4.24VAVNVPIVNPFNFDD310 pKa = 3.56GEE312 pKa = 4.4DD313 pKa = 3.37SGRR316 pKa = 11.84EE317 pKa = 3.86SSQTGGNANQPPATKK332 pKa = 10.0RR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84QKK337 pKa = 10.73SRR339 pKa = 11.84AEE341 pKa = 3.92RR342 pKa = 11.84EE343 pKa = 4.2AEE345 pKa = 3.9RR346 pKa = 11.84LQIDD350 pKa = 3.77QPVRR354 pKa = 11.84TQARR358 pKa = 11.84TSRR361 pKa = 11.84VHH363 pKa = 5.9NKK365 pKa = 9.3SRR367 pKa = 11.84NAGGQDD373 pKa = 2.87QDD375 pKa = 3.82LQIVGVGAGEE385 pKa = 4.81DD386 pKa = 3.72DD387 pKa = 3.76PHH389 pKa = 7.84VQEE392 pKa = 4.79QVV394 pKa = 2.81
MM1 pKa = 7.95ADD3 pKa = 4.38FEE5 pKa = 4.39AQRR8 pKa = 11.84DD9 pKa = 3.92DD10 pKa = 5.06NIAANAPIGGLSSKK24 pKa = 8.01QQEE27 pKa = 4.29IDD29 pKa = 3.4TVTAAITPLLAAGFPQAIFNYY50 pKa = 10.32DD51 pKa = 3.5DD52 pKa = 4.24LLLRR56 pKa = 11.84GYY58 pKa = 7.12TAKK61 pKa = 10.03TFCDD65 pKa = 5.23FIKK68 pKa = 10.16PLSAITEE75 pKa = 4.04RR76 pKa = 11.84RR77 pKa = 11.84EE78 pKa = 4.35LISLCALGNNRR89 pKa = 11.84FWDD92 pKa = 3.6MSVVAEE98 pKa = 4.57LDD100 pKa = 3.35EE101 pKa = 4.34FLNFVKK107 pKa = 10.02WLKK110 pKa = 10.79SPEE113 pKa = 3.9GRR115 pKa = 11.84DD116 pKa = 3.51AQTQAAKK123 pKa = 10.32KK124 pKa = 9.67RR125 pKa = 11.84ALNKK129 pKa = 9.82KK130 pKa = 9.92ASDD133 pKa = 3.5GMSTKK138 pKa = 10.57DD139 pKa = 3.32VALVQMGNAIIADD152 pKa = 3.74YY153 pKa = 10.58QRR155 pKa = 11.84EE156 pKa = 4.18RR157 pKa = 11.84KK158 pKa = 8.4QRR160 pKa = 11.84RR161 pKa = 11.84FPIEE165 pKa = 3.64EE166 pKa = 3.69EE167 pKa = 3.84MAEE170 pKa = 4.25LRR172 pKa = 11.84RR173 pKa = 11.84QLRR176 pKa = 11.84QLDD179 pKa = 4.0EE180 pKa = 4.07EE181 pKa = 4.34LQAVEE186 pKa = 4.6EE187 pKa = 4.45EE188 pKa = 4.43IKK190 pKa = 11.01VKK192 pKa = 10.49YY193 pKa = 10.57GPVALYY199 pKa = 8.86EE200 pKa = 4.64GPDD203 pKa = 3.4NTRR206 pKa = 11.84VKK208 pKa = 10.78SDD210 pKa = 3.59AYY212 pKa = 11.43LMYY215 pKa = 10.52QDD217 pKa = 4.71DD218 pKa = 4.42CRR220 pKa = 11.84KK221 pKa = 9.74KK222 pKa = 9.78GYY224 pKa = 9.7RR225 pKa = 11.84AIAQYY230 pKa = 10.71QGGFEE235 pKa = 4.14KK236 pKa = 10.62AVEE239 pKa = 3.94LFGNKK244 pKa = 7.29VRR246 pKa = 11.84EE247 pKa = 4.21AHH249 pKa = 5.71FCAYY253 pKa = 10.48LSDD256 pKa = 4.01PARR259 pKa = 11.84SDD261 pKa = 3.67FVRR264 pKa = 11.84DD265 pKa = 3.53YY266 pKa = 9.79YY267 pKa = 10.07NQKK270 pKa = 9.46IVHH273 pKa = 6.41LEE275 pKa = 3.55RR276 pKa = 11.84SGEE279 pKa = 4.08KK280 pKa = 9.92KK281 pKa = 9.73QAGSFRR287 pKa = 11.84SLLVAAHH294 pKa = 6.63GEE296 pKa = 4.24VAVNVPIVNPFNFDD310 pKa = 3.56GEE312 pKa = 4.4DD313 pKa = 3.37SGRR316 pKa = 11.84EE317 pKa = 3.86SSQTGGNANQPPATKK332 pKa = 10.0RR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84QKK337 pKa = 10.73SRR339 pKa = 11.84AEE341 pKa = 3.92RR342 pKa = 11.84EE343 pKa = 4.2AEE345 pKa = 3.9RR346 pKa = 11.84LQIDD350 pKa = 3.77QPVRR354 pKa = 11.84TQARR358 pKa = 11.84TSRR361 pKa = 11.84VHH363 pKa = 5.9NKK365 pKa = 9.3SRR367 pKa = 11.84NAGGQDD373 pKa = 2.87QDD375 pKa = 3.82LQIVGVGAGEE385 pKa = 4.81DD386 pKa = 3.72DD387 pKa = 3.76PHH389 pKa = 7.84VQEE392 pKa = 4.79QVV394 pKa = 2.81
Molecular weight: 44.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V0A0B8|A0A2V0A0B8_9VIRU ORF1+2p OS=Medicago sativa amalgavirus 1 OX=2069326 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.95ADD3 pKa = 4.38FEE5 pKa = 4.39AQRR8 pKa = 11.84DD9 pKa = 3.92DD10 pKa = 5.06NIAANAPIGGLSSKK24 pKa = 8.01QQEE27 pKa = 4.29IDD29 pKa = 3.4TVTAAITPLLAAGFPQAIFNYY50 pKa = 10.32DD51 pKa = 3.5DD52 pKa = 4.24LLLRR56 pKa = 11.84GYY58 pKa = 7.12TAKK61 pKa = 10.03TFCDD65 pKa = 5.23FIKK68 pKa = 10.16PLSAITEE75 pKa = 4.04RR76 pKa = 11.84RR77 pKa = 11.84EE78 pKa = 4.35LISLCALGNNRR89 pKa = 11.84FWDD92 pKa = 3.6MSVVAEE98 pKa = 4.57LDD100 pKa = 3.35EE101 pKa = 4.34FLNFVKK107 pKa = 10.02WLKK110 pKa = 10.79SPEE113 pKa = 3.9GRR115 pKa = 11.84DD116 pKa = 3.51AQTQAAKK123 pKa = 10.32KK124 pKa = 9.67RR125 pKa = 11.84ALNKK129 pKa = 9.82KK130 pKa = 9.92ASDD133 pKa = 3.5GMSTKK138 pKa = 10.57DD139 pKa = 3.32VALVQMGNAIIADD152 pKa = 3.74YY153 pKa = 10.58QRR155 pKa = 11.84EE156 pKa = 4.18RR157 pKa = 11.84KK158 pKa = 8.4QRR160 pKa = 11.84RR161 pKa = 11.84FPIEE165 pKa = 3.64EE166 pKa = 3.69EE167 pKa = 3.84MAEE170 pKa = 4.25LRR172 pKa = 11.84RR173 pKa = 11.84QLRR176 pKa = 11.84QLDD179 pKa = 4.0EE180 pKa = 4.07EE181 pKa = 4.34LQAVEE186 pKa = 4.6EE187 pKa = 4.45EE188 pKa = 4.43IKK190 pKa = 11.01VKK192 pKa = 10.49YY193 pKa = 10.57GPVALYY199 pKa = 8.86EE200 pKa = 4.64GPDD203 pKa = 3.4NTRR206 pKa = 11.84VKK208 pKa = 10.78SDD210 pKa = 3.59AYY212 pKa = 11.43LMYY215 pKa = 10.52QDD217 pKa = 4.71DD218 pKa = 4.42CRR220 pKa = 11.84KK221 pKa = 9.74KK222 pKa = 9.78GYY224 pKa = 9.7RR225 pKa = 11.84AIAQYY230 pKa = 10.71QGGFEE235 pKa = 4.14KK236 pKa = 10.62AVEE239 pKa = 3.94LFGNKK244 pKa = 7.29VRR246 pKa = 11.84EE247 pKa = 4.21AHH249 pKa = 5.71FCAYY253 pKa = 10.48LSDD256 pKa = 4.01PARR259 pKa = 11.84SDD261 pKa = 3.67FVRR264 pKa = 11.84DD265 pKa = 3.53YY266 pKa = 9.79YY267 pKa = 10.07NQKK270 pKa = 9.46IVHH273 pKa = 6.41LEE275 pKa = 3.55RR276 pKa = 11.84SGEE279 pKa = 4.08KK280 pKa = 9.98KK281 pKa = 9.91QAGSFAVSWWPLMEE295 pKa = 4.54KK296 pKa = 9.44WLLMFPLSTRR306 pKa = 11.84STLMEE311 pKa = 5.3KK312 pKa = 10.09IPVGKK317 pKa = 8.72VHH319 pKa = 7.07KK320 pKa = 10.31PGEE323 pKa = 4.46TPTNRR328 pKa = 11.84PLLRR332 pKa = 11.84GADD335 pKa = 3.42KK336 pKa = 11.25SRR338 pKa = 11.84VLNVRR343 pKa = 11.84QSGYY347 pKa = 10.79KK348 pKa = 10.01SISRR352 pKa = 11.84LEE354 pKa = 4.25RR355 pKa = 11.84KK356 pKa = 9.69HH357 pKa = 6.23EE358 pKa = 3.98LQGCITSQGMPEE370 pKa = 3.72GRR372 pKa = 11.84IKK374 pKa = 10.4IFKK377 pKa = 9.66SWEE380 pKa = 4.05SEE382 pKa = 4.0PGRR385 pKa = 11.84MIPMSRR391 pKa = 11.84SKK393 pKa = 11.12YY394 pKa = 7.64EE395 pKa = 3.86CAVRR399 pKa = 11.84WVIGGGEE406 pKa = 3.96VRR408 pKa = 11.84NWKK411 pKa = 9.66VDD413 pKa = 2.82SSMYY417 pKa = 10.35RR418 pKa = 11.84GGGSSNDD425 pKa = 3.5ALKK428 pKa = 11.08LLANASTVRR437 pKa = 11.84PGKK440 pKa = 10.0LLRR443 pKa = 11.84DD444 pKa = 3.67VYY446 pKa = 10.47SFRR449 pKa = 11.84VARR452 pKa = 11.84ARR454 pKa = 11.84LQLPGDD460 pKa = 4.01FSVPDD465 pKa = 4.08GPDD468 pKa = 2.69ACRR471 pKa = 11.84VKK473 pKa = 10.94NFNNFATSGPVLKK486 pKa = 10.94AFGVRR491 pKa = 11.84SKK493 pKa = 11.42NGLRR497 pKa = 11.84QLLQDD502 pKa = 3.67EE503 pKa = 4.49AWWYY507 pKa = 10.52FNSFGNSDD515 pKa = 3.93FGVEE519 pKa = 3.94GLPWFGARR527 pKa = 11.84LGFRR531 pKa = 11.84SKK533 pKa = 10.97LVTEE537 pKa = 4.58EE538 pKa = 3.97KK539 pKa = 10.41ARR541 pKa = 11.84KK542 pKa = 9.2KK543 pKa = 10.28ISEE546 pKa = 4.15GDD548 pKa = 3.75SVGRR552 pKa = 11.84AVMMMDD558 pKa = 5.17ALEE561 pKa = 4.23QCCSSPLYY569 pKa = 10.51NVLSTYY575 pKa = 8.45TFHH578 pKa = 7.56KK579 pKa = 10.07RR580 pKa = 11.84LNRR583 pKa = 11.84RR584 pKa = 11.84SGFKK588 pKa = 10.34NAVVKK593 pKa = 10.89ASSDD597 pKa = 3.05WAHH600 pKa = 6.24IWNGVKK606 pKa = 10.23DD607 pKa = 3.51AAVIVEE613 pKa = 4.4LDD615 pKa = 2.89WSKK618 pKa = 10.98FDD620 pKa = 3.95RR621 pKa = 11.84EE622 pKa = 4.19RR623 pKa = 11.84PRR625 pKa = 11.84EE626 pKa = 3.97DD627 pKa = 3.66LEE629 pKa = 4.65FMVSLISSCFNPKK642 pKa = 9.27SARR645 pKa = 11.84EE646 pKa = 3.8EE647 pKa = 4.22RR648 pKa = 11.84LLHH651 pKa = 7.2AYY653 pKa = 5.92TTSNFRR659 pKa = 11.84ALVEE663 pKa = 4.16RR664 pKa = 11.84PVFLDD669 pKa = 3.17GGGVFGIEE677 pKa = 3.97GMVPSGSLWTGWLDD691 pKa = 3.31TALNILYY698 pKa = 8.53MKK700 pKa = 10.05AVCAEE705 pKa = 4.06IGVGDD710 pKa = 4.56DD711 pKa = 4.32DD712 pKa = 5.55VEE714 pKa = 4.64VMCAGDD720 pKa = 4.8DD721 pKa = 3.67NLTLFKK727 pKa = 10.59FDD729 pKa = 4.01PGEE732 pKa = 4.06ANLRR736 pKa = 11.84RR737 pKa = 11.84IRR739 pKa = 11.84DD740 pKa = 3.74LLNDD744 pKa = 3.7WFLAGISEE752 pKa = 4.74DD753 pKa = 3.12EE754 pKa = 4.4FLIHH758 pKa = 6.97RR759 pKa = 11.84PPYY762 pKa = 10.28HH763 pKa = 4.66VTKK766 pKa = 10.7VQACFPEE773 pKa = 4.8GTDD776 pKa = 3.77LSRR779 pKa = 11.84GTSGMLKK786 pKa = 9.64KK787 pKa = 10.59AVWVPFEE794 pKa = 4.32GEE796 pKa = 4.29VVIDD800 pKa = 3.58NARR803 pKa = 11.84GRR805 pKa = 11.84SHH807 pKa = 6.44RR808 pKa = 11.84WEE810 pKa = 4.06YY811 pKa = 10.99RR812 pKa = 11.84FSGCPKK818 pKa = 10.15FLSCYY823 pKa = 8.11WLHH826 pKa = 7.32DD827 pKa = 3.8GLPIRR832 pKa = 11.84PAHH835 pKa = 6.99DD836 pKa = 3.77NLEE839 pKa = 4.15KK840 pKa = 10.97LLWPEE845 pKa = 4.36GLHH848 pKa = 7.51DD849 pKa = 6.67DD850 pKa = 3.4IDD852 pKa = 4.38VYY854 pKa = 10.95EE855 pKa = 4.41GAVISMVVDD864 pKa = 3.69NPHH867 pKa = 5.86NHH869 pKa = 6.37HH870 pKa = 6.75NCNHH874 pKa = 5.61MLSRR878 pKa = 11.84FIIIRR883 pKa = 11.84EE884 pKa = 3.93ARR886 pKa = 11.84RR887 pKa = 11.84LGASVEE893 pKa = 4.39DD894 pKa = 3.7PFIPIKK900 pKa = 10.37HH901 pKa = 5.49GSIRR905 pKa = 11.84PVGDD909 pKa = 3.26EE910 pKa = 3.72PVPYY914 pKa = 10.02PEE916 pKa = 4.13LAPWRR921 pKa = 11.84RR922 pKa = 11.84APGGYY927 pKa = 9.84KK928 pKa = 10.4LEE930 pKa = 5.09DD931 pKa = 3.48YY932 pKa = 10.68PEE934 pKa = 4.29NAEE937 pKa = 4.42HH938 pKa = 6.15IQVFRR943 pKa = 11.84DD944 pKa = 3.97FMQGVSSLYY953 pKa = 10.32LRR955 pKa = 11.84EE956 pKa = 4.05ATGGVDD962 pKa = 2.83AWQFMDD968 pKa = 5.25IIRR971 pKa = 11.84GDD973 pKa = 3.53AFVGEE978 pKa = 4.47GQFGNDD984 pKa = 3.17LRR986 pKa = 11.84SWLNWLHH993 pKa = 5.94HH994 pKa = 6.34HH995 pKa = 7.41PISKK999 pKa = 10.09YY1000 pKa = 10.17LRR1002 pKa = 11.84EE1003 pKa = 4.18TKK1005 pKa = 9.23TFRR1008 pKa = 11.84LKK1010 pKa = 10.98DD1011 pKa = 3.52GADD1014 pKa = 3.3PDD1016 pKa = 4.0APKK1019 pKa = 10.72DD1020 pKa = 3.63LEE1022 pKa = 4.2KK1023 pKa = 10.86AGRR1026 pKa = 11.84ALRR1029 pKa = 11.84ALRR1032 pKa = 11.84GRR1034 pKa = 11.84LEE1036 pKa = 4.34LGGFACVEE1044 pKa = 4.13DD1045 pKa = 4.49FVRR1048 pKa = 11.84WLDD1051 pKa = 4.05GIRR1054 pKa = 11.84SSRR1057 pKa = 11.84EE1058 pKa = 3.58
MM1 pKa = 7.95ADD3 pKa = 4.38FEE5 pKa = 4.39AQRR8 pKa = 11.84DD9 pKa = 3.92DD10 pKa = 5.06NIAANAPIGGLSSKK24 pKa = 8.01QQEE27 pKa = 4.29IDD29 pKa = 3.4TVTAAITPLLAAGFPQAIFNYY50 pKa = 10.32DD51 pKa = 3.5DD52 pKa = 4.24LLLRR56 pKa = 11.84GYY58 pKa = 7.12TAKK61 pKa = 10.03TFCDD65 pKa = 5.23FIKK68 pKa = 10.16PLSAITEE75 pKa = 4.04RR76 pKa = 11.84RR77 pKa = 11.84EE78 pKa = 4.35LISLCALGNNRR89 pKa = 11.84FWDD92 pKa = 3.6MSVVAEE98 pKa = 4.57LDD100 pKa = 3.35EE101 pKa = 4.34FLNFVKK107 pKa = 10.02WLKK110 pKa = 10.79SPEE113 pKa = 3.9GRR115 pKa = 11.84DD116 pKa = 3.51AQTQAAKK123 pKa = 10.32KK124 pKa = 9.67RR125 pKa = 11.84ALNKK129 pKa = 9.82KK130 pKa = 9.92ASDD133 pKa = 3.5GMSTKK138 pKa = 10.57DD139 pKa = 3.32VALVQMGNAIIADD152 pKa = 3.74YY153 pKa = 10.58QRR155 pKa = 11.84EE156 pKa = 4.18RR157 pKa = 11.84KK158 pKa = 8.4QRR160 pKa = 11.84RR161 pKa = 11.84FPIEE165 pKa = 3.64EE166 pKa = 3.69EE167 pKa = 3.84MAEE170 pKa = 4.25LRR172 pKa = 11.84RR173 pKa = 11.84QLRR176 pKa = 11.84QLDD179 pKa = 4.0EE180 pKa = 4.07EE181 pKa = 4.34LQAVEE186 pKa = 4.6EE187 pKa = 4.45EE188 pKa = 4.43IKK190 pKa = 11.01VKK192 pKa = 10.49YY193 pKa = 10.57GPVALYY199 pKa = 8.86EE200 pKa = 4.64GPDD203 pKa = 3.4NTRR206 pKa = 11.84VKK208 pKa = 10.78SDD210 pKa = 3.59AYY212 pKa = 11.43LMYY215 pKa = 10.52QDD217 pKa = 4.71DD218 pKa = 4.42CRR220 pKa = 11.84KK221 pKa = 9.74KK222 pKa = 9.78GYY224 pKa = 9.7RR225 pKa = 11.84AIAQYY230 pKa = 10.71QGGFEE235 pKa = 4.14KK236 pKa = 10.62AVEE239 pKa = 3.94LFGNKK244 pKa = 7.29VRR246 pKa = 11.84EE247 pKa = 4.21AHH249 pKa = 5.71FCAYY253 pKa = 10.48LSDD256 pKa = 4.01PARR259 pKa = 11.84SDD261 pKa = 3.67FVRR264 pKa = 11.84DD265 pKa = 3.53YY266 pKa = 9.79YY267 pKa = 10.07NQKK270 pKa = 9.46IVHH273 pKa = 6.41LEE275 pKa = 3.55RR276 pKa = 11.84SGEE279 pKa = 4.08KK280 pKa = 9.98KK281 pKa = 9.91QAGSFAVSWWPLMEE295 pKa = 4.54KK296 pKa = 9.44WLLMFPLSTRR306 pKa = 11.84STLMEE311 pKa = 5.3KK312 pKa = 10.09IPVGKK317 pKa = 8.72VHH319 pKa = 7.07KK320 pKa = 10.31PGEE323 pKa = 4.46TPTNRR328 pKa = 11.84PLLRR332 pKa = 11.84GADD335 pKa = 3.42KK336 pKa = 11.25SRR338 pKa = 11.84VLNVRR343 pKa = 11.84QSGYY347 pKa = 10.79KK348 pKa = 10.01SISRR352 pKa = 11.84LEE354 pKa = 4.25RR355 pKa = 11.84KK356 pKa = 9.69HH357 pKa = 6.23EE358 pKa = 3.98LQGCITSQGMPEE370 pKa = 3.72GRR372 pKa = 11.84IKK374 pKa = 10.4IFKK377 pKa = 9.66SWEE380 pKa = 4.05SEE382 pKa = 4.0PGRR385 pKa = 11.84MIPMSRR391 pKa = 11.84SKK393 pKa = 11.12YY394 pKa = 7.64EE395 pKa = 3.86CAVRR399 pKa = 11.84WVIGGGEE406 pKa = 3.96VRR408 pKa = 11.84NWKK411 pKa = 9.66VDD413 pKa = 2.82SSMYY417 pKa = 10.35RR418 pKa = 11.84GGGSSNDD425 pKa = 3.5ALKK428 pKa = 11.08LLANASTVRR437 pKa = 11.84PGKK440 pKa = 10.0LLRR443 pKa = 11.84DD444 pKa = 3.67VYY446 pKa = 10.47SFRR449 pKa = 11.84VARR452 pKa = 11.84ARR454 pKa = 11.84LQLPGDD460 pKa = 4.01FSVPDD465 pKa = 4.08GPDD468 pKa = 2.69ACRR471 pKa = 11.84VKK473 pKa = 10.94NFNNFATSGPVLKK486 pKa = 10.94AFGVRR491 pKa = 11.84SKK493 pKa = 11.42NGLRR497 pKa = 11.84QLLQDD502 pKa = 3.67EE503 pKa = 4.49AWWYY507 pKa = 10.52FNSFGNSDD515 pKa = 3.93FGVEE519 pKa = 3.94GLPWFGARR527 pKa = 11.84LGFRR531 pKa = 11.84SKK533 pKa = 10.97LVTEE537 pKa = 4.58EE538 pKa = 3.97KK539 pKa = 10.41ARR541 pKa = 11.84KK542 pKa = 9.2KK543 pKa = 10.28ISEE546 pKa = 4.15GDD548 pKa = 3.75SVGRR552 pKa = 11.84AVMMMDD558 pKa = 5.17ALEE561 pKa = 4.23QCCSSPLYY569 pKa = 10.51NVLSTYY575 pKa = 8.45TFHH578 pKa = 7.56KK579 pKa = 10.07RR580 pKa = 11.84LNRR583 pKa = 11.84RR584 pKa = 11.84SGFKK588 pKa = 10.34NAVVKK593 pKa = 10.89ASSDD597 pKa = 3.05WAHH600 pKa = 6.24IWNGVKK606 pKa = 10.23DD607 pKa = 3.51AAVIVEE613 pKa = 4.4LDD615 pKa = 2.89WSKK618 pKa = 10.98FDD620 pKa = 3.95RR621 pKa = 11.84EE622 pKa = 4.19RR623 pKa = 11.84PRR625 pKa = 11.84EE626 pKa = 3.97DD627 pKa = 3.66LEE629 pKa = 4.65FMVSLISSCFNPKK642 pKa = 9.27SARR645 pKa = 11.84EE646 pKa = 3.8EE647 pKa = 4.22RR648 pKa = 11.84LLHH651 pKa = 7.2AYY653 pKa = 5.92TTSNFRR659 pKa = 11.84ALVEE663 pKa = 4.16RR664 pKa = 11.84PVFLDD669 pKa = 3.17GGGVFGIEE677 pKa = 3.97GMVPSGSLWTGWLDD691 pKa = 3.31TALNILYY698 pKa = 8.53MKK700 pKa = 10.05AVCAEE705 pKa = 4.06IGVGDD710 pKa = 4.56DD711 pKa = 4.32DD712 pKa = 5.55VEE714 pKa = 4.64VMCAGDD720 pKa = 4.8DD721 pKa = 3.67NLTLFKK727 pKa = 10.59FDD729 pKa = 4.01PGEE732 pKa = 4.06ANLRR736 pKa = 11.84RR737 pKa = 11.84IRR739 pKa = 11.84DD740 pKa = 3.74LLNDD744 pKa = 3.7WFLAGISEE752 pKa = 4.74DD753 pKa = 3.12EE754 pKa = 4.4FLIHH758 pKa = 6.97RR759 pKa = 11.84PPYY762 pKa = 10.28HH763 pKa = 4.66VTKK766 pKa = 10.7VQACFPEE773 pKa = 4.8GTDD776 pKa = 3.77LSRR779 pKa = 11.84GTSGMLKK786 pKa = 9.64KK787 pKa = 10.59AVWVPFEE794 pKa = 4.32GEE796 pKa = 4.29VVIDD800 pKa = 3.58NARR803 pKa = 11.84GRR805 pKa = 11.84SHH807 pKa = 6.44RR808 pKa = 11.84WEE810 pKa = 4.06YY811 pKa = 10.99RR812 pKa = 11.84FSGCPKK818 pKa = 10.15FLSCYY823 pKa = 8.11WLHH826 pKa = 7.32DD827 pKa = 3.8GLPIRR832 pKa = 11.84PAHH835 pKa = 6.99DD836 pKa = 3.77NLEE839 pKa = 4.15KK840 pKa = 10.97LLWPEE845 pKa = 4.36GLHH848 pKa = 7.51DD849 pKa = 6.67DD850 pKa = 3.4IDD852 pKa = 4.38VYY854 pKa = 10.95EE855 pKa = 4.41GAVISMVVDD864 pKa = 3.69NPHH867 pKa = 5.86NHH869 pKa = 6.37HH870 pKa = 6.75NCNHH874 pKa = 5.61MLSRR878 pKa = 11.84FIIIRR883 pKa = 11.84EE884 pKa = 3.93ARR886 pKa = 11.84RR887 pKa = 11.84LGASVEE893 pKa = 4.39DD894 pKa = 3.7PFIPIKK900 pKa = 10.37HH901 pKa = 5.49GSIRR905 pKa = 11.84PVGDD909 pKa = 3.26EE910 pKa = 3.72PVPYY914 pKa = 10.02PEE916 pKa = 4.13LAPWRR921 pKa = 11.84RR922 pKa = 11.84APGGYY927 pKa = 9.84KK928 pKa = 10.4LEE930 pKa = 5.09DD931 pKa = 3.48YY932 pKa = 10.68PEE934 pKa = 4.29NAEE937 pKa = 4.42HH938 pKa = 6.15IQVFRR943 pKa = 11.84DD944 pKa = 3.97FMQGVSSLYY953 pKa = 10.32LRR955 pKa = 11.84EE956 pKa = 4.05ATGGVDD962 pKa = 2.83AWQFMDD968 pKa = 5.25IIRR971 pKa = 11.84GDD973 pKa = 3.53AFVGEE978 pKa = 4.47GQFGNDD984 pKa = 3.17LRR986 pKa = 11.84SWLNWLHH993 pKa = 5.94HH994 pKa = 6.34HH995 pKa = 7.41PISKK999 pKa = 10.09YY1000 pKa = 10.17LRR1002 pKa = 11.84EE1003 pKa = 4.18TKK1005 pKa = 9.23TFRR1008 pKa = 11.84LKK1010 pKa = 10.98DD1011 pKa = 3.52GADD1014 pKa = 3.3PDD1016 pKa = 4.0APKK1019 pKa = 10.72DD1020 pKa = 3.63LEE1022 pKa = 4.2KK1023 pKa = 10.86AGRR1026 pKa = 11.84ALRR1029 pKa = 11.84ALRR1032 pKa = 11.84GRR1034 pKa = 11.84LEE1036 pKa = 4.34LGGFACVEE1044 pKa = 4.13DD1045 pKa = 4.49FVRR1048 pKa = 11.84WLDD1051 pKa = 4.05GIRR1054 pKa = 11.84SSRR1057 pKa = 11.84EE1058 pKa = 3.58
Molecular weight: 120.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1452 |
394 |
1058 |
726.0 |
82.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.609 ± 1.223 | 1.446 ± 0.229 |
6.749 ± 0.19 | 7.163 ± 0.374 |
4.683 ± 0.33 | 7.576 ± 0.518 |
1.86 ± 0.313 | 4.339 ± 0.013 |
5.854 ± 0.126 | 8.54 ± 0.76 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.325 | 4.063 ± 0.268 |
4.545 ± 0.392 | 3.926 ± 1.687 |
8.196 ± 0.096 | 6.129 ± 0.559 |
2.961 ± 0.179 | 6.405 ± 0.032 |
1.928 ± 0.754 | 2.893 ± 0.081 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
