
Muricauda alvinocaridis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Muricauda
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
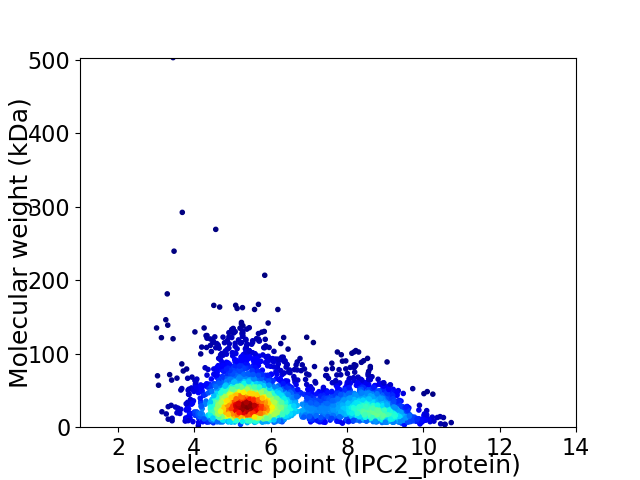
Virtual 2D-PAGE plot for 3339 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S8RQJ5|A0A4S8RQJ5_9FLAO Uncharacterized protein OS=Muricauda alvinocaridis OX=2530200 GN=EZV76_15095 PE=4 SV=1
MM1 pKa = 6.73NTYY4 pKa = 10.41ILRR7 pKa = 11.84LLLLCMLFLCAPSCNQDD24 pKa = 3.24DD25 pKa = 4.43TVVDD29 pKa = 4.55DD30 pKa = 5.92IINEE34 pKa = 4.17EE35 pKa = 4.62PDD37 pKa = 3.95PDD39 pKa = 4.76PNPDD43 pKa = 4.46PDD45 pKa = 5.19PDD47 pKa = 4.95PDD49 pKa = 4.97PDD51 pKa = 3.49TDD53 pKa = 3.82KK54 pKa = 11.28PIFEE58 pKa = 5.27GDD60 pKa = 3.14TGNVLVYY67 pKa = 10.6DD68 pKa = 3.96EE69 pKa = 5.16EE70 pKa = 5.77KK71 pKa = 10.81IFDD74 pKa = 4.19GYY76 pKa = 11.58VLVNDD81 pKa = 4.09ASDD84 pKa = 3.36NRR86 pKa = 11.84VYY88 pKa = 11.57LMTKK92 pKa = 8.82EE93 pKa = 3.95GKK95 pKa = 9.01IRR97 pKa = 11.84YY98 pKa = 7.55EE99 pKa = 3.65WEE101 pKa = 3.84LPSNIGNDD109 pKa = 3.63AEE111 pKa = 4.19LLEE114 pKa = 4.32NGKK117 pKa = 9.46LLVALTDD124 pKa = 3.41EE125 pKa = 4.2NAFYY129 pKa = 11.0DD130 pKa = 3.4VGGFGGRR137 pKa = 11.84IQIINPDD144 pKa = 3.74RR145 pKa = 11.84SVEE148 pKa = 3.7WDD150 pKa = 3.52FLYY153 pKa = 9.91STEE156 pKa = 5.35DD157 pKa = 3.8YY158 pKa = 10.51ISHH161 pKa = 7.15HH162 pKa = 7.38DD163 pKa = 4.57IEE165 pKa = 4.73MLPNGNVLILAWQKK179 pKa = 10.7RR180 pKa = 11.84SKK182 pKa = 10.84EE183 pKa = 4.0DD184 pKa = 3.44ALQAGYY190 pKa = 10.83DD191 pKa = 4.04GIPDD195 pKa = 3.74NEE197 pKa = 4.2LLLLEE202 pKa = 4.6SLIEE206 pKa = 4.12VNPEE210 pKa = 3.47NNDD213 pKa = 3.7IVWEE217 pKa = 4.06WHH219 pKa = 5.82SWDD222 pKa = 5.68HH223 pKa = 7.29IIQDD227 pKa = 3.89FDD229 pKa = 3.61DD230 pKa = 4.37TKK232 pKa = 11.68DD233 pKa = 3.44SFGDD237 pKa = 3.28IAEE240 pKa = 4.3NPQLIDD246 pKa = 5.12LNFHH250 pKa = 7.19DD251 pKa = 5.84DD252 pKa = 3.58DD253 pKa = 6.66RR254 pKa = 11.84GDD256 pKa = 3.33IMHH259 pKa = 7.31ANGLDD264 pKa = 3.64YY265 pKa = 11.37DD266 pKa = 4.2PVNDD270 pKa = 5.89LIYY273 pKa = 10.76LSVNFYY279 pKa = 11.12SEE281 pKa = 3.91VWVIDD286 pKa = 3.46HH287 pKa = 6.2STTTEE292 pKa = 3.66EE293 pKa = 4.12AATNSGGNYY302 pKa = 9.88NKK304 pKa = 10.79GGDD307 pKa = 3.43LVYY310 pKa = 10.85RR311 pKa = 11.84FGNPLAYY318 pKa = 9.63KK319 pKa = 10.4NEE321 pKa = 4.02TGTRR325 pKa = 11.84LFYY328 pKa = 11.12NNHH331 pKa = 5.8FPNYY335 pKa = 9.53LDD337 pKa = 3.57SDD339 pKa = 4.28EE340 pKa = 5.2IGAGNLLIYY349 pKa = 10.43MNGNNGSQQQSHH361 pKa = 5.75VYY363 pKa = 9.57EE364 pKa = 4.58LNLPDD369 pKa = 5.39SFTLLPEE376 pKa = 4.34NDD378 pKa = 3.79NEE380 pKa = 4.21PTIEE384 pKa = 4.06WEE386 pKa = 4.11FTDD389 pKa = 4.32AEE391 pKa = 4.65LYY393 pKa = 10.8SPIVSGAVRR402 pKa = 11.84LPNGNTLIAEE412 pKa = 4.11GRR414 pKa = 11.84YY415 pKa = 9.91GYY417 pKa = 10.09WEE419 pKa = 4.01VTEE422 pKa = 4.23SGEE425 pKa = 4.62VVWLFEE431 pKa = 4.32GNKK434 pKa = 9.96LFWRR438 pKa = 11.84GYY440 pKa = 9.99SYY442 pKa = 11.23PKK444 pKa = 10.06DD445 pKa = 3.61HH446 pKa = 7.29PGVLLLEE453 pKa = 4.76LNN455 pKa = 3.91
MM1 pKa = 6.73NTYY4 pKa = 10.41ILRR7 pKa = 11.84LLLLCMLFLCAPSCNQDD24 pKa = 3.24DD25 pKa = 4.43TVVDD29 pKa = 4.55DD30 pKa = 5.92IINEE34 pKa = 4.17EE35 pKa = 4.62PDD37 pKa = 3.95PDD39 pKa = 4.76PNPDD43 pKa = 4.46PDD45 pKa = 5.19PDD47 pKa = 4.95PDD49 pKa = 4.97PDD51 pKa = 3.49TDD53 pKa = 3.82KK54 pKa = 11.28PIFEE58 pKa = 5.27GDD60 pKa = 3.14TGNVLVYY67 pKa = 10.6DD68 pKa = 3.96EE69 pKa = 5.16EE70 pKa = 5.77KK71 pKa = 10.81IFDD74 pKa = 4.19GYY76 pKa = 11.58VLVNDD81 pKa = 4.09ASDD84 pKa = 3.36NRR86 pKa = 11.84VYY88 pKa = 11.57LMTKK92 pKa = 8.82EE93 pKa = 3.95GKK95 pKa = 9.01IRR97 pKa = 11.84YY98 pKa = 7.55EE99 pKa = 3.65WEE101 pKa = 3.84LPSNIGNDD109 pKa = 3.63AEE111 pKa = 4.19LLEE114 pKa = 4.32NGKK117 pKa = 9.46LLVALTDD124 pKa = 3.41EE125 pKa = 4.2NAFYY129 pKa = 11.0DD130 pKa = 3.4VGGFGGRR137 pKa = 11.84IQIINPDD144 pKa = 3.74RR145 pKa = 11.84SVEE148 pKa = 3.7WDD150 pKa = 3.52FLYY153 pKa = 9.91STEE156 pKa = 5.35DD157 pKa = 3.8YY158 pKa = 10.51ISHH161 pKa = 7.15HH162 pKa = 7.38DD163 pKa = 4.57IEE165 pKa = 4.73MLPNGNVLILAWQKK179 pKa = 10.7RR180 pKa = 11.84SKK182 pKa = 10.84EE183 pKa = 4.0DD184 pKa = 3.44ALQAGYY190 pKa = 10.83DD191 pKa = 4.04GIPDD195 pKa = 3.74NEE197 pKa = 4.2LLLLEE202 pKa = 4.6SLIEE206 pKa = 4.12VNPEE210 pKa = 3.47NNDD213 pKa = 3.7IVWEE217 pKa = 4.06WHH219 pKa = 5.82SWDD222 pKa = 5.68HH223 pKa = 7.29IIQDD227 pKa = 3.89FDD229 pKa = 3.61DD230 pKa = 4.37TKK232 pKa = 11.68DD233 pKa = 3.44SFGDD237 pKa = 3.28IAEE240 pKa = 4.3NPQLIDD246 pKa = 5.12LNFHH250 pKa = 7.19DD251 pKa = 5.84DD252 pKa = 3.58DD253 pKa = 6.66RR254 pKa = 11.84GDD256 pKa = 3.33IMHH259 pKa = 7.31ANGLDD264 pKa = 3.64YY265 pKa = 11.37DD266 pKa = 4.2PVNDD270 pKa = 5.89LIYY273 pKa = 10.76LSVNFYY279 pKa = 11.12SEE281 pKa = 3.91VWVIDD286 pKa = 3.46HH287 pKa = 6.2STTTEE292 pKa = 3.66EE293 pKa = 4.12AATNSGGNYY302 pKa = 9.88NKK304 pKa = 10.79GGDD307 pKa = 3.43LVYY310 pKa = 10.85RR311 pKa = 11.84FGNPLAYY318 pKa = 9.63KK319 pKa = 10.4NEE321 pKa = 4.02TGTRR325 pKa = 11.84LFYY328 pKa = 11.12NNHH331 pKa = 5.8FPNYY335 pKa = 9.53LDD337 pKa = 3.57SDD339 pKa = 4.28EE340 pKa = 5.2IGAGNLLIYY349 pKa = 10.43MNGNNGSQQQSHH361 pKa = 5.75VYY363 pKa = 9.57EE364 pKa = 4.58LNLPDD369 pKa = 5.39SFTLLPEE376 pKa = 4.34NDD378 pKa = 3.79NEE380 pKa = 4.21PTIEE384 pKa = 4.06WEE386 pKa = 4.11FTDD389 pKa = 4.32AEE391 pKa = 4.65LYY393 pKa = 10.8SPIVSGAVRR402 pKa = 11.84LPNGNTLIAEE412 pKa = 4.11GRR414 pKa = 11.84YY415 pKa = 9.91GYY417 pKa = 10.09WEE419 pKa = 4.01VTEE422 pKa = 4.23SGEE425 pKa = 4.62VVWLFEE431 pKa = 4.32GNKK434 pKa = 9.96LFWRR438 pKa = 11.84GYY440 pKa = 9.99SYY442 pKa = 11.23PKK444 pKa = 10.06DD445 pKa = 3.61HH446 pKa = 7.29PGVLLLEE453 pKa = 4.76LNN455 pKa = 3.91
Molecular weight: 51.79 kDa
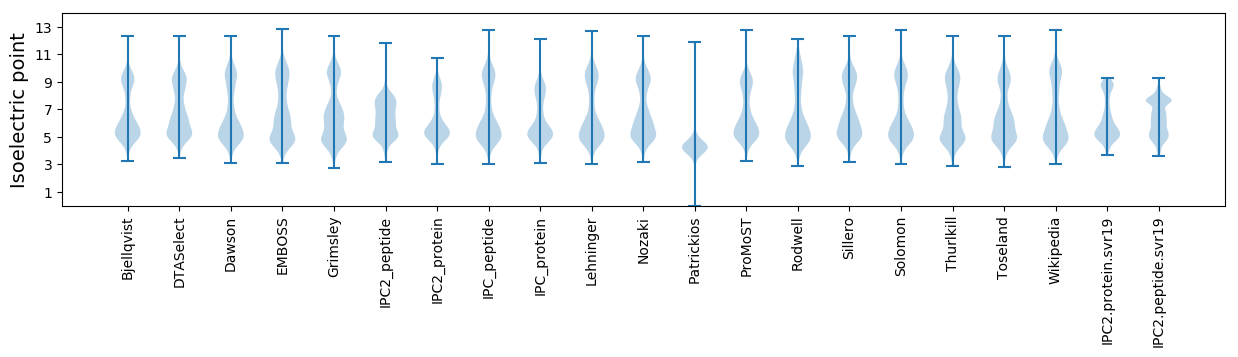
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S8RUP1|A0A4S8RUP1_9FLAO Ferredoxin--NADP reductase OS=Muricauda alvinocaridis OX=2530200 GN=EZV76_04575 PE=3 SV=1
MM1 pKa = 7.75KK2 pKa = 10.5YY3 pKa = 10.47SPLWRR8 pKa = 11.84ILWSRR13 pKa = 11.84FRR15 pKa = 11.84FPKK18 pKa = 10.0MPLQLRR24 pKa = 11.84SGVWLKK30 pKa = 10.67IPPICHH36 pKa = 6.4FDD38 pKa = 3.6MRR40 pKa = 11.84CGTLTEE46 pKa = 4.92KK47 pKa = 10.39SHH49 pKa = 5.99QLIISHH55 pKa = 6.29SSFGYY60 pKa = 9.13SQDD63 pKa = 3.48RR64 pKa = 11.84ARR66 pKa = 11.84NDD68 pKa = 3.27KK69 pKa = 10.81KK70 pKa = 10.9NSKK73 pKa = 9.94
MM1 pKa = 7.75KK2 pKa = 10.5YY3 pKa = 10.47SPLWRR8 pKa = 11.84ILWSRR13 pKa = 11.84FRR15 pKa = 11.84FPKK18 pKa = 10.0MPLQLRR24 pKa = 11.84SGVWLKK30 pKa = 10.67IPPICHH36 pKa = 6.4FDD38 pKa = 3.6MRR40 pKa = 11.84CGTLTEE46 pKa = 4.92KK47 pKa = 10.39SHH49 pKa = 5.99QLIISHH55 pKa = 6.29SSFGYY60 pKa = 9.13SQDD63 pKa = 3.48RR64 pKa = 11.84ARR66 pKa = 11.84NDD68 pKa = 3.27KK69 pKa = 10.81KK70 pKa = 10.9NSKK73 pKa = 9.94
Molecular weight: 8.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1128126 |
18 |
4754 |
337.9 |
38.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.581 ± 0.043 | 0.744 ± 0.014 |
5.758 ± 0.039 | 6.841 ± 0.039 |
5.101 ± 0.032 | 6.987 ± 0.042 |
1.959 ± 0.024 | 7.085 ± 0.039 |
7.129 ± 0.059 | 9.485 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.436 ± 0.025 | 5.395 ± 0.043 |
3.66 ± 0.028 | 3.599 ± 0.023 |
3.773 ± 0.031 | 6.272 ± 0.034 |
5.614 ± 0.051 | 6.56 ± 0.032 |
1.136 ± 0.018 | 3.869 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
