
Mycobacterium conspicuum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
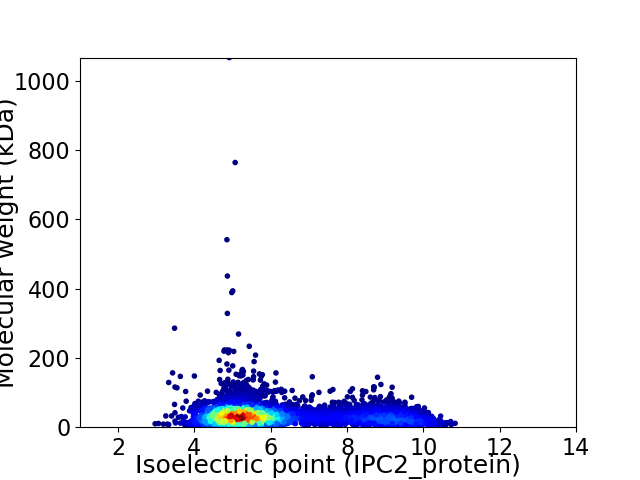
Virtual 2D-PAGE plot for 5457 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X1T608|A0A1X1T608_9MYCO Transglutaminase OS=Mycobacterium conspicuum OX=44010 GN=AWC00_17595 PE=4 SV=1
MM1 pKa = 7.36SFVIAASDD9 pKa = 3.3AMAAATTDD17 pKa = 3.81LADD20 pKa = 4.3LGSTLRR26 pKa = 11.84SAHH29 pKa = 6.07AVAAPATTGLVAAGADD45 pKa = 3.58EE46 pKa = 4.49VSAAVATLFGSHH58 pKa = 5.68AQAYY62 pKa = 7.36QAISAHH68 pKa = 5.57AAEE71 pKa = 4.29FHH73 pKa = 6.74SRR75 pKa = 11.84FVQLLSAGAGQYY87 pKa = 11.68ALTEE91 pKa = 4.15ATNASPLQAVEE102 pKa = 4.57HH103 pKa = 6.29GLLNAINAPTQALVGRR119 pKa = 11.84PLIGNGADD127 pKa = 3.46GVAGGTLAQANGGAGGILFGNGGAGATDD155 pKa = 3.41AAGQGGSGGAAGLIGNGGAGGAGATGGAGGRR186 pKa = 11.84GGAGGLLYY194 pKa = 11.04GNGGAGGSGGDD205 pKa = 3.94STDD208 pKa = 3.34GAAGGAGGAGGSAGFFGNGGAGGSGGAGGDD238 pKa = 3.35ATVAGGNGGAGGNGGAGGNAGHH260 pKa = 6.49VLGDD264 pKa = 4.03GGAGGAGGAGGGGGAVTGGTVISGDD289 pKa = 2.97IGGAGGNGGAGGPTFVGQGGNGGAGGAGGAAGAVTDD325 pKa = 3.73STIYY329 pKa = 10.95SGDD332 pKa = 3.28TGGAGGNGGAGNLAGLFGNGGHH354 pKa = 6.75GGAGGNGGAAGTVTNSTVQTGDD376 pKa = 2.77TGGAGGAGGAASLGGFVGNGGNGGAGGAGGDD407 pKa = 3.57AGAVTGGSNIATGDD421 pKa = 3.41NGGAGGDD428 pKa = 3.44GGAGNIAAYY437 pKa = 10.08VGSGGNGGAGGNGGNAGAVSASSTVDD463 pKa = 3.01NGDD466 pKa = 3.32NGGDD470 pKa = 3.7GGDD473 pKa = 3.87GGTGTGGASSFGGTASHH490 pKa = 6.95GGNGGAGGAAGSVTDD505 pKa = 4.05SAVHH509 pKa = 6.3SGDD512 pKa = 3.3NGGAGGDD519 pKa = 3.59GGTIVAGKK527 pKa = 9.78GGNGGNGGNGGDD539 pKa = 3.86GGALTGSTVTSGDD552 pKa = 3.24NGGTGGAGGAGGMIGTSGNGGNGGDD577 pKa = 4.2GGDD580 pKa = 3.84GGAVSASSAVTVGDD594 pKa = 3.4NGGAGGAGGAGSSSGYY610 pKa = 9.1FANGGNGGAGGNGGAGGAVSGSSTVSTGDD639 pKa = 3.01NGGAGGAGGAGAIAGFIGNGGNGGAGGDD667 pKa = 3.68GGGGGGG673 pKa = 3.88
MM1 pKa = 7.36SFVIAASDD9 pKa = 3.3AMAAATTDD17 pKa = 3.81LADD20 pKa = 4.3LGSTLRR26 pKa = 11.84SAHH29 pKa = 6.07AVAAPATTGLVAAGADD45 pKa = 3.58EE46 pKa = 4.49VSAAVATLFGSHH58 pKa = 5.68AQAYY62 pKa = 7.36QAISAHH68 pKa = 5.57AAEE71 pKa = 4.29FHH73 pKa = 6.74SRR75 pKa = 11.84FVQLLSAGAGQYY87 pKa = 11.68ALTEE91 pKa = 4.15ATNASPLQAVEE102 pKa = 4.57HH103 pKa = 6.29GLLNAINAPTQALVGRR119 pKa = 11.84PLIGNGADD127 pKa = 3.46GVAGGTLAQANGGAGGILFGNGGAGATDD155 pKa = 3.41AAGQGGSGGAAGLIGNGGAGGAGATGGAGGRR186 pKa = 11.84GGAGGLLYY194 pKa = 11.04GNGGAGGSGGDD205 pKa = 3.94STDD208 pKa = 3.34GAAGGAGGAGGSAGFFGNGGAGGSGGAGGDD238 pKa = 3.35ATVAGGNGGAGGNGGAGGNAGHH260 pKa = 6.49VLGDD264 pKa = 4.03GGAGGAGGAGGGGGAVTGGTVISGDD289 pKa = 2.97IGGAGGNGGAGGPTFVGQGGNGGAGGAGGAAGAVTDD325 pKa = 3.73STIYY329 pKa = 10.95SGDD332 pKa = 3.28TGGAGGNGGAGNLAGLFGNGGHH354 pKa = 6.75GGAGGNGGAAGTVTNSTVQTGDD376 pKa = 2.77TGGAGGAGGAASLGGFVGNGGNGGAGGAGGDD407 pKa = 3.57AGAVTGGSNIATGDD421 pKa = 3.41NGGAGGDD428 pKa = 3.44GGAGNIAAYY437 pKa = 10.08VGSGGNGGAGGNGGNAGAVSASSTVDD463 pKa = 3.01NGDD466 pKa = 3.32NGGDD470 pKa = 3.7GGDD473 pKa = 3.87GGTGTGGASSFGGTASHH490 pKa = 6.95GGNGGAGGAAGSVTDD505 pKa = 4.05SAVHH509 pKa = 6.3SGDD512 pKa = 3.3NGGAGGDD519 pKa = 3.59GGTIVAGKK527 pKa = 9.78GGNGGNGGNGGDD539 pKa = 3.86GGALTGSTVTSGDD552 pKa = 3.24NGGTGGAGGAGGMIGTSGNGGNGGDD577 pKa = 4.2GGDD580 pKa = 3.84GGAVSASSAVTVGDD594 pKa = 3.4NGGAGGAGGAGSSSGYY610 pKa = 9.1FANGGNGGAGGNGGAGGAVSGSSTVSTGDD639 pKa = 3.01NGGAGGAGGAGAIAGFIGNGGNGGAGGDD667 pKa = 3.68GGGGGGG673 pKa = 3.88
Molecular weight: 55.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X1T242|A0A1X1T242_9MYCO Alanine and proline-rich secreted protein Apa OS=Mycobacterium conspicuum OX=44010 GN=AWC00_19620 PE=3 SV=1
MM1 pKa = 7.61PGITRR6 pKa = 11.84SSAHH10 pKa = 5.54STSSIAEE17 pKa = 4.1RR18 pKa = 11.84LTLRR22 pKa = 11.84PDD24 pKa = 3.4TTRR27 pKa = 11.84TASRR31 pKa = 11.84NTDD34 pKa = 2.81GAADD38 pKa = 3.86AGRR41 pKa = 11.84PRR43 pKa = 11.84VPEE46 pKa = 4.09AGKK49 pKa = 10.19AAGRR53 pKa = 11.84EE54 pKa = 4.28SGSGRR59 pKa = 11.84SDD61 pKa = 2.95GGSGVDD67 pKa = 3.72RR68 pKa = 11.84VPAGTNGLVICAKK81 pKa = 9.42SVRR84 pKa = 11.84DD85 pKa = 3.77TVDD88 pKa = 4.03GIVDD92 pKa = 3.93AEE94 pKa = 4.4LSASDD99 pKa = 3.81ALGLRR104 pKa = 11.84FPRR107 pKa = 11.84PNMVASAAALFEE119 pKa = 4.39PAEE122 pKa = 4.22PWAHH126 pKa = 5.62GRR128 pKa = 11.84DD129 pKa = 3.66RR130 pKa = 11.84LDD132 pKa = 3.37VEE134 pKa = 4.61PEE136 pKa = 3.81CGRR139 pKa = 11.84FRR141 pKa = 11.84ITIGPGVVRR150 pKa = 11.84LGWTNPVRR158 pKa = 11.84AEE160 pKa = 4.02KK161 pKa = 10.56ASEE164 pKa = 4.09RR165 pKa = 11.84TVGQHH170 pKa = 4.8MQDD173 pKa = 2.94VAGEE177 pKa = 3.99VDD179 pKa = 4.09RR180 pKa = 11.84IKK182 pKa = 10.86AGRR185 pKa = 11.84DD186 pKa = 3.27VPDD189 pKa = 3.89PSGRR193 pKa = 11.84AITEE197 pKa = 3.83WSRR200 pKa = 11.84KK201 pKa = 8.11SRR203 pKa = 11.84AAMCRR208 pKa = 11.84TFAEE212 pKa = 4.0LDD214 pKa = 3.51YY215 pKa = 11.07TPMVEE220 pKa = 4.21SGRR223 pKa = 11.84VPAMVTLTYY232 pKa = 10.39PGDD235 pKa = 3.94WEE237 pKa = 4.76SVAPDD242 pKa = 3.41GASVKK247 pKa = 10.04RR248 pKa = 11.84HH249 pKa = 3.72MMLWRR254 pKa = 11.84KK255 pKa = 9.49RR256 pKa = 11.84FQRR259 pKa = 11.84EE260 pKa = 3.24WGEE263 pKa = 3.53PARR266 pKa = 11.84YY267 pKa = 7.08IWKK270 pKa = 8.8MEE272 pKa = 3.95FQRR275 pKa = 11.84RR276 pKa = 11.84GAPHH280 pKa = 5.72IHH282 pKa = 6.48LWMAPPYY289 pKa = 8.9TVGRR293 pKa = 11.84SGRR296 pKa = 11.84RR297 pKa = 11.84FSEE300 pKa = 3.94WLSQEE305 pKa = 3.45WAEE308 pKa = 4.63VVAHH312 pKa = 7.05PDD314 pKa = 3.09PEE316 pKa = 3.96QRR318 pKa = 11.84TRR320 pKa = 11.84HH321 pKa = 5.3LRR323 pKa = 11.84AGTAIDD329 pKa = 3.61SLNGLRR335 pKa = 11.84ACDD338 pKa = 3.92PKK340 pKa = 10.89RR341 pKa = 11.84LAIYY345 pKa = 7.5FTKK348 pKa = 10.43HH349 pKa = 5.28SSPNRR354 pKa = 11.84LGDD357 pKa = 3.4KK358 pKa = 9.68EE359 pKa = 4.21YY360 pKa = 10.91QHH362 pKa = 6.79IVPEE366 pKa = 4.14AWRR369 pKa = 11.84RR370 pKa = 11.84PGRR373 pKa = 11.84GPGRR377 pKa = 11.84FWGVHH382 pKa = 3.94GLEE385 pKa = 4.08RR386 pKa = 11.84ATAVVEE392 pKa = 4.31IAQDD396 pKa = 3.18AYY398 pKa = 9.93LTARR402 pKa = 11.84RR403 pKa = 11.84IIRR406 pKa = 11.84RR407 pKa = 11.84WSRR410 pKa = 11.84SQAIHH415 pKa = 6.74SDD417 pKa = 3.57CTHH420 pKa = 7.07CFPTALVPRR429 pKa = 11.84MTRR432 pKa = 11.84VAVQRR437 pKa = 11.84TNSKK441 pKa = 7.97TGKK444 pKa = 8.26VRR446 pKa = 11.84HH447 pKa = 6.12RR448 pKa = 11.84NVCRR452 pKa = 11.84RR453 pKa = 11.84RR454 pKa = 11.84LLCRR458 pKa = 11.84QGGFAGGYY466 pKa = 10.28ALVNDD471 pKa = 5.09GPAFASQLAAALSADD486 pKa = 3.75RR487 pKa = 11.84AKK489 pKa = 10.73AVRR492 pKa = 11.84DD493 pKa = 3.44EE494 pKa = 4.02QLAGRR499 pKa = 11.84KK500 pKa = 8.49
MM1 pKa = 7.61PGITRR6 pKa = 11.84SSAHH10 pKa = 5.54STSSIAEE17 pKa = 4.1RR18 pKa = 11.84LTLRR22 pKa = 11.84PDD24 pKa = 3.4TTRR27 pKa = 11.84TASRR31 pKa = 11.84NTDD34 pKa = 2.81GAADD38 pKa = 3.86AGRR41 pKa = 11.84PRR43 pKa = 11.84VPEE46 pKa = 4.09AGKK49 pKa = 10.19AAGRR53 pKa = 11.84EE54 pKa = 4.28SGSGRR59 pKa = 11.84SDD61 pKa = 2.95GGSGVDD67 pKa = 3.72RR68 pKa = 11.84VPAGTNGLVICAKK81 pKa = 9.42SVRR84 pKa = 11.84DD85 pKa = 3.77TVDD88 pKa = 4.03GIVDD92 pKa = 3.93AEE94 pKa = 4.4LSASDD99 pKa = 3.81ALGLRR104 pKa = 11.84FPRR107 pKa = 11.84PNMVASAAALFEE119 pKa = 4.39PAEE122 pKa = 4.22PWAHH126 pKa = 5.62GRR128 pKa = 11.84DD129 pKa = 3.66RR130 pKa = 11.84LDD132 pKa = 3.37VEE134 pKa = 4.61PEE136 pKa = 3.81CGRR139 pKa = 11.84FRR141 pKa = 11.84ITIGPGVVRR150 pKa = 11.84LGWTNPVRR158 pKa = 11.84AEE160 pKa = 4.02KK161 pKa = 10.56ASEE164 pKa = 4.09RR165 pKa = 11.84TVGQHH170 pKa = 4.8MQDD173 pKa = 2.94VAGEE177 pKa = 3.99VDD179 pKa = 4.09RR180 pKa = 11.84IKK182 pKa = 10.86AGRR185 pKa = 11.84DD186 pKa = 3.27VPDD189 pKa = 3.89PSGRR193 pKa = 11.84AITEE197 pKa = 3.83WSRR200 pKa = 11.84KK201 pKa = 8.11SRR203 pKa = 11.84AAMCRR208 pKa = 11.84TFAEE212 pKa = 4.0LDD214 pKa = 3.51YY215 pKa = 11.07TPMVEE220 pKa = 4.21SGRR223 pKa = 11.84VPAMVTLTYY232 pKa = 10.39PGDD235 pKa = 3.94WEE237 pKa = 4.76SVAPDD242 pKa = 3.41GASVKK247 pKa = 10.04RR248 pKa = 11.84HH249 pKa = 3.72MMLWRR254 pKa = 11.84KK255 pKa = 9.49RR256 pKa = 11.84FQRR259 pKa = 11.84EE260 pKa = 3.24WGEE263 pKa = 3.53PARR266 pKa = 11.84YY267 pKa = 7.08IWKK270 pKa = 8.8MEE272 pKa = 3.95FQRR275 pKa = 11.84RR276 pKa = 11.84GAPHH280 pKa = 5.72IHH282 pKa = 6.48LWMAPPYY289 pKa = 8.9TVGRR293 pKa = 11.84SGRR296 pKa = 11.84RR297 pKa = 11.84FSEE300 pKa = 3.94WLSQEE305 pKa = 3.45WAEE308 pKa = 4.63VVAHH312 pKa = 7.05PDD314 pKa = 3.09PEE316 pKa = 3.96QRR318 pKa = 11.84TRR320 pKa = 11.84HH321 pKa = 5.3LRR323 pKa = 11.84AGTAIDD329 pKa = 3.61SLNGLRR335 pKa = 11.84ACDD338 pKa = 3.92PKK340 pKa = 10.89RR341 pKa = 11.84LAIYY345 pKa = 7.5FTKK348 pKa = 10.43HH349 pKa = 5.28SSPNRR354 pKa = 11.84LGDD357 pKa = 3.4KK358 pKa = 9.68EE359 pKa = 4.21YY360 pKa = 10.91QHH362 pKa = 6.79IVPEE366 pKa = 4.14AWRR369 pKa = 11.84RR370 pKa = 11.84PGRR373 pKa = 11.84GPGRR377 pKa = 11.84FWGVHH382 pKa = 3.94GLEE385 pKa = 4.08RR386 pKa = 11.84ATAVVEE392 pKa = 4.31IAQDD396 pKa = 3.18AYY398 pKa = 9.93LTARR402 pKa = 11.84RR403 pKa = 11.84IIRR406 pKa = 11.84RR407 pKa = 11.84WSRR410 pKa = 11.84SQAIHH415 pKa = 6.74SDD417 pKa = 3.57CTHH420 pKa = 7.07CFPTALVPRR429 pKa = 11.84MTRR432 pKa = 11.84VAVQRR437 pKa = 11.84TNSKK441 pKa = 7.97TGKK444 pKa = 8.26VRR446 pKa = 11.84HH447 pKa = 6.12RR448 pKa = 11.84NVCRR452 pKa = 11.84RR453 pKa = 11.84RR454 pKa = 11.84LLCRR458 pKa = 11.84QGGFAGGYY466 pKa = 10.28ALVNDD471 pKa = 5.09GPAFASQLAAALSADD486 pKa = 3.75RR487 pKa = 11.84AKK489 pKa = 10.73AVRR492 pKa = 11.84DD493 pKa = 3.44EE494 pKa = 4.02QLAGRR499 pKa = 11.84KK500 pKa = 8.49
Molecular weight: 55.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1826921 |
42 |
10008 |
334.8 |
35.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.137 ± 0.049 | 0.834 ± 0.01 |
6.189 ± 0.027 | 5.1 ± 0.036 |
3.081 ± 0.019 | 9.442 ± 0.145 |
2.256 ± 0.015 | 4.241 ± 0.023 |
2.195 ± 0.022 | 9.738 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.03 ± 0.015 | 2.438 ± 0.032 |
5.831 ± 0.033 | 3.082 ± 0.015 |
7.166 ± 0.041 | 5.369 ± 0.023 |
5.828 ± 0.026 | 8.381 ± 0.035 |
1.496 ± 0.013 | 2.167 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
