
Rice latent virus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.78
Get precalculated fractions of proteins
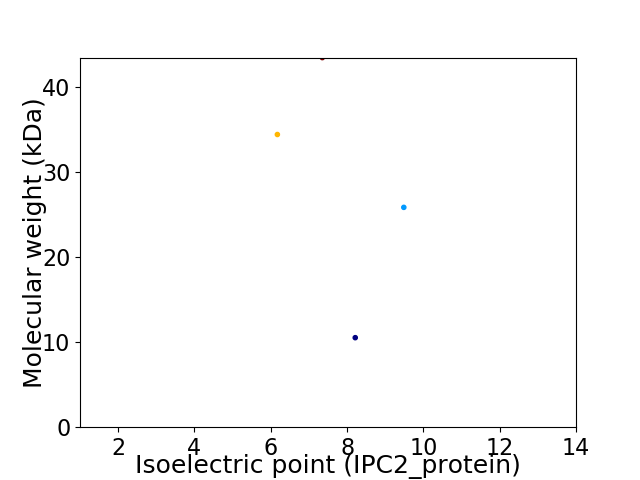
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D0WZF4|A0A2D0WZF4_9GEMI Replication-associated protein OS=Rice latent virus 2 OX=2012857 PE=3 SV=1
MM1 pKa = 7.63TNTEE5 pKa = 4.18SASEE9 pKa = 4.06QPHH12 pKa = 6.03NFAFQNKK19 pKa = 7.79NVFLTYY25 pKa = 9.47PRR27 pKa = 11.84CSQEE31 pKa = 3.66PHH33 pKa = 5.46TMGLFLWNLLRR44 pKa = 11.84PYY46 pKa = 10.81GVLCASEE53 pKa = 4.11LHH55 pKa = 6.94KK56 pKa = 11.21DD57 pKa = 3.9GTPHH61 pKa = 6.9LHH63 pKa = 6.99ALAQTRR69 pKa = 11.84HH70 pKa = 6.97RR71 pKa = 11.84IRR73 pKa = 11.84THH75 pKa = 6.75DD76 pKa = 3.52SSFFDD81 pKa = 4.11NMDD84 pKa = 3.44YY85 pKa = 10.92HH86 pKa = 7.07PNIQGTRR93 pKa = 11.84NPKK96 pKa = 10.15AVLAYY101 pKa = 9.52ILKK104 pKa = 9.97HH105 pKa = 5.53PTGEE109 pKa = 3.62WHH111 pKa = 6.77KK112 pKa = 11.24GSFRR116 pKa = 11.84PRR118 pKa = 11.84GSFGVPPTGNGEE130 pKa = 4.32AGPSTTIRR138 pKa = 11.84RR139 pKa = 11.84AAGDD143 pKa = 3.64SSSSAEE149 pKa = 4.04GGNTSSEE156 pKa = 4.2LANKK160 pKa = 9.02RR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84MTKK166 pKa = 9.73DD167 pKa = 2.84QIMADD172 pKa = 3.59ILRR175 pKa = 11.84SATSRR180 pKa = 11.84MEE182 pKa = 3.79YY183 pKa = 9.9MNGVKK188 pKa = 9.68KK189 pKa = 9.99WFPYY193 pKa = 9.99DD194 pKa = 3.23YY195 pKa = 10.39CARR198 pKa = 11.84LQAWEE203 pKa = 4.19YY204 pKa = 11.12ASQKK208 pKa = 10.26LFPDD212 pKa = 3.79PPVTYY217 pKa = 9.67EE218 pKa = 3.92PPFNDD223 pKa = 4.38SMFHH227 pKa = 5.2CHH229 pKa = 5.44EE230 pKa = 4.78TLTEE234 pKa = 3.83WAAEE238 pKa = 4.21NIYY241 pKa = 10.68HH242 pKa = 5.66VTPQVYY248 pKa = 10.36SLLHH252 pKa = 5.19PHH254 pKa = 6.63SDD256 pKa = 3.47AEE258 pKa = 4.5ADD260 pKa = 4.22LRR262 pKa = 11.84WLHH265 pKa = 6.0DD266 pKa = 3.09TCFASRR272 pKa = 11.84RR273 pKa = 11.84DD274 pKa = 3.71QSASTSADD282 pKa = 3.16HH283 pKa = 6.88QGQEE287 pKa = 4.15SLLGLEE293 pKa = 4.9ASADD297 pKa = 3.99TITTRR302 pKa = 11.84ITT304 pKa = 3.03
MM1 pKa = 7.63TNTEE5 pKa = 4.18SASEE9 pKa = 4.06QPHH12 pKa = 6.03NFAFQNKK19 pKa = 7.79NVFLTYY25 pKa = 9.47PRR27 pKa = 11.84CSQEE31 pKa = 3.66PHH33 pKa = 5.46TMGLFLWNLLRR44 pKa = 11.84PYY46 pKa = 10.81GVLCASEE53 pKa = 4.11LHH55 pKa = 6.94KK56 pKa = 11.21DD57 pKa = 3.9GTPHH61 pKa = 6.9LHH63 pKa = 6.99ALAQTRR69 pKa = 11.84HH70 pKa = 6.97RR71 pKa = 11.84IRR73 pKa = 11.84THH75 pKa = 6.75DD76 pKa = 3.52SSFFDD81 pKa = 4.11NMDD84 pKa = 3.44YY85 pKa = 10.92HH86 pKa = 7.07PNIQGTRR93 pKa = 11.84NPKK96 pKa = 10.15AVLAYY101 pKa = 9.52ILKK104 pKa = 9.97HH105 pKa = 5.53PTGEE109 pKa = 3.62WHH111 pKa = 6.77KK112 pKa = 11.24GSFRR116 pKa = 11.84PRR118 pKa = 11.84GSFGVPPTGNGEE130 pKa = 4.32AGPSTTIRR138 pKa = 11.84RR139 pKa = 11.84AAGDD143 pKa = 3.64SSSSAEE149 pKa = 4.04GGNTSSEE156 pKa = 4.2LANKK160 pKa = 9.02RR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84MTKK166 pKa = 9.73DD167 pKa = 2.84QIMADD172 pKa = 3.59ILRR175 pKa = 11.84SATSRR180 pKa = 11.84MEE182 pKa = 3.79YY183 pKa = 9.9MNGVKK188 pKa = 9.68KK189 pKa = 9.99WFPYY193 pKa = 9.99DD194 pKa = 3.23YY195 pKa = 10.39CARR198 pKa = 11.84LQAWEE203 pKa = 4.19YY204 pKa = 11.12ASQKK208 pKa = 10.26LFPDD212 pKa = 3.79PPVTYY217 pKa = 9.67EE218 pKa = 3.92PPFNDD223 pKa = 4.38SMFHH227 pKa = 5.2CHH229 pKa = 5.44EE230 pKa = 4.78TLTEE234 pKa = 3.83WAAEE238 pKa = 4.21NIYY241 pKa = 10.68HH242 pKa = 5.66VTPQVYY248 pKa = 10.36SLLHH252 pKa = 5.19PHH254 pKa = 6.63SDD256 pKa = 3.47AEE258 pKa = 4.5ADD260 pKa = 4.22LRR262 pKa = 11.84WLHH265 pKa = 6.0DD266 pKa = 3.09TCFASRR272 pKa = 11.84RR273 pKa = 11.84DD274 pKa = 3.71QSASTSADD282 pKa = 3.16HH283 pKa = 6.88QGQEE287 pKa = 4.15SLLGLEE293 pKa = 4.9ASADD297 pKa = 3.99TITTRR302 pKa = 11.84ITT304 pKa = 3.03
Molecular weight: 34.37 kDa
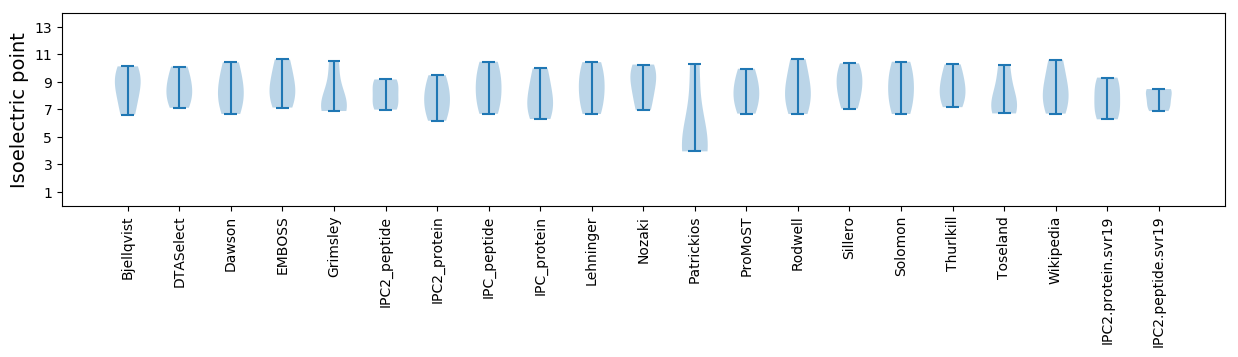
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D0WZE5|A0A2D0WZE5_9GEMI Movement protein OS=Rice latent virus 2 OX=2012857 PE=3 SV=1
MM1 pKa = 7.63PARR4 pKa = 11.84TGSARR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 10.35RR12 pKa = 11.84KK13 pKa = 9.87DD14 pKa = 2.87DD15 pKa = 3.82WAWDD19 pKa = 3.32SATRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84VRR27 pKa = 11.84PRR29 pKa = 11.84TSYY32 pKa = 10.63VPRR35 pKa = 11.84GGPRR39 pKa = 11.84VRR41 pKa = 11.84RR42 pKa = 11.84PSLQYY47 pKa = 11.11VNPGLLGSTAVHH59 pKa = 5.78VADD62 pKa = 3.92TNGEE66 pKa = 3.93VRR68 pKa = 11.84IITGYY73 pKa = 10.68SRR75 pKa = 11.84GSDD78 pKa = 3.21EE79 pKa = 4.66QSRR82 pKa = 11.84HH83 pKa = 4.17TNEE86 pKa = 3.98TITYY90 pKa = 9.21KK91 pKa = 10.82LSLDD95 pKa = 3.79LYY97 pKa = 8.16VTVDD101 pKa = 3.7TEE103 pKa = 4.02CQKK106 pKa = 11.1YY107 pKa = 8.98VGKK110 pKa = 10.43GVAVAWLVYY119 pKa = 9.79DD120 pKa = 4.19AQPTGEE126 pKa = 4.46MPAASTIFPHH136 pKa = 5.72VADD139 pKa = 4.57MATAPCTWKK148 pKa = 10.46VGRR151 pKa = 11.84EE152 pKa = 3.84VCHH155 pKa = 6.21RR156 pKa = 11.84FVVKK160 pKa = 10.3RR161 pKa = 11.84RR162 pKa = 11.84WVITLEE168 pKa = 4.21TNGRR172 pKa = 11.84VAGTVFQGANGVPPCNRR189 pKa = 11.84TVYY192 pKa = 9.79FHH194 pKa = 7.13KK195 pKa = 9.97FCKK198 pKa = 10.03RR199 pKa = 11.84LGVRR203 pKa = 11.84TEE205 pKa = 3.77WKK207 pKa = 7.92NTSGGSIGDD216 pKa = 3.7IKK218 pKa = 10.2TGAPVHH224 pKa = 5.44SVRR227 pKa = 11.84GRR229 pKa = 11.84EE230 pKa = 3.75PDD232 pKa = 2.99
MM1 pKa = 7.63PARR4 pKa = 11.84TGSARR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 10.35RR12 pKa = 11.84KK13 pKa = 9.87DD14 pKa = 2.87DD15 pKa = 3.82WAWDD19 pKa = 3.32SATRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84VRR27 pKa = 11.84PRR29 pKa = 11.84TSYY32 pKa = 10.63VPRR35 pKa = 11.84GGPRR39 pKa = 11.84VRR41 pKa = 11.84RR42 pKa = 11.84PSLQYY47 pKa = 11.11VNPGLLGSTAVHH59 pKa = 5.78VADD62 pKa = 3.92TNGEE66 pKa = 3.93VRR68 pKa = 11.84IITGYY73 pKa = 10.68SRR75 pKa = 11.84GSDD78 pKa = 3.21EE79 pKa = 4.66QSRR82 pKa = 11.84HH83 pKa = 4.17TNEE86 pKa = 3.98TITYY90 pKa = 9.21KK91 pKa = 10.82LSLDD95 pKa = 3.79LYY97 pKa = 8.16VTVDD101 pKa = 3.7TEE103 pKa = 4.02CQKK106 pKa = 11.1YY107 pKa = 8.98VGKK110 pKa = 10.43GVAVAWLVYY119 pKa = 9.79DD120 pKa = 4.19AQPTGEE126 pKa = 4.46MPAASTIFPHH136 pKa = 5.72VADD139 pKa = 4.57MATAPCTWKK148 pKa = 10.46VGRR151 pKa = 11.84EE152 pKa = 3.84VCHH155 pKa = 6.21RR156 pKa = 11.84FVVKK160 pKa = 10.3RR161 pKa = 11.84RR162 pKa = 11.84WVITLEE168 pKa = 4.21TNGRR172 pKa = 11.84VAGTVFQGANGVPPCNRR189 pKa = 11.84TVYY192 pKa = 9.79FHH194 pKa = 7.13KK195 pKa = 9.97FCKK198 pKa = 10.03RR199 pKa = 11.84LGVRR203 pKa = 11.84TEE205 pKa = 3.77WKK207 pKa = 7.92NTSGGSIGDD216 pKa = 3.7IKK218 pKa = 10.2TGAPVHH224 pKa = 5.44SVRR227 pKa = 11.84GRR229 pKa = 11.84EE230 pKa = 3.75PDD232 pKa = 2.99
Molecular weight: 25.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1016 |
98 |
382 |
254.0 |
28.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.874 ± 0.367 | 2.264 ± 0.289 |
4.823 ± 0.417 | 4.626 ± 0.424 |
4.035 ± 0.592 | 7.283 ± 1.04 |
3.74 ± 0.748 | 3.642 ± 0.555 |
4.331 ± 0.488 | 6.398 ± 0.935 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.461 ± 0.348 | 4.429 ± 0.816 |
6.693 ± 0.285 | 3.445 ± 0.53 |
7.185 ± 1.221 | 7.48 ± 0.692 |
7.874 ± 0.604 | 5.315 ± 2.027 |
2.264 ± 0.178 | 3.839 ± 0.559 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
