
gamma proteobacterium HIMB55
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; unclassified Halieaceae
Average proteome isoelectric point is 5.59
Get precalculated fractions of proteins
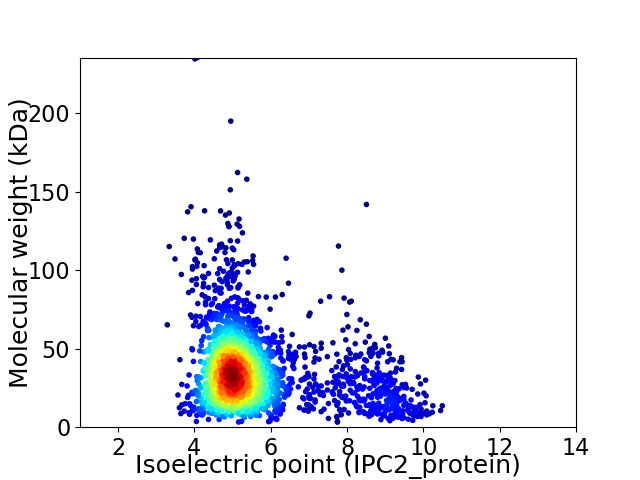
Virtual 2D-PAGE plot for 2470 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
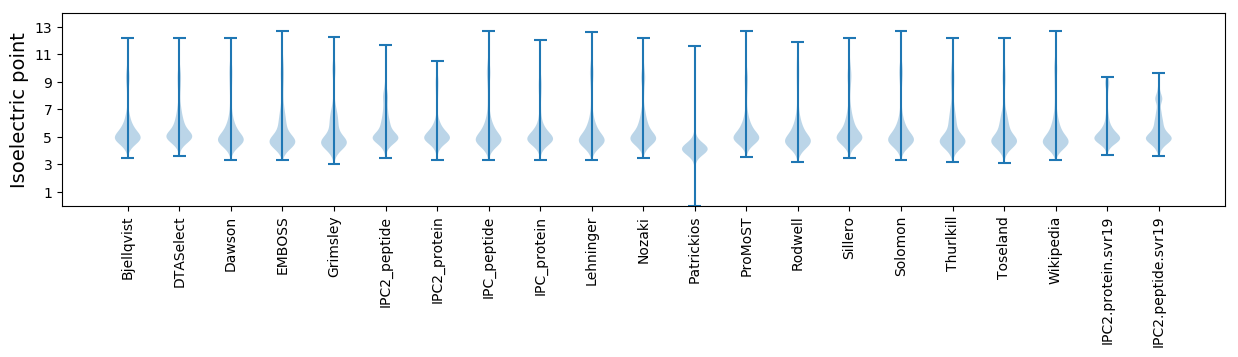
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H3NY45|H3NY45_9GAMM Amino acid carrier protein OS=gamma proteobacterium HIMB55 OX=745014 GN=OMB55_00004860 PE=3 SV=1
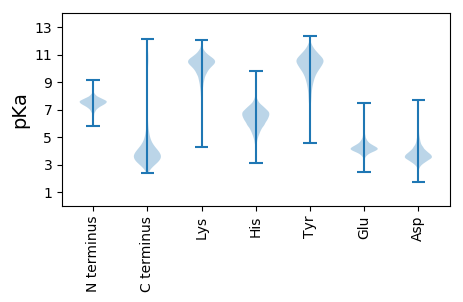
MM1 pKa = 7.04NRR3 pKa = 11.84APVFDD8 pKa = 3.74QASYY12 pKa = 11.1ALSVQEE18 pKa = 4.13NTGTTISGISFSDD31 pKa = 3.67PDD33 pKa = 4.34ADD35 pKa = 5.47PITLSISGTDD45 pKa = 3.33SEE47 pKa = 4.86AFSVTSIGSLAFITPPDD64 pKa = 3.91FEE66 pKa = 4.83TPADD70 pKa = 3.49SDD72 pKa = 3.8QNNEE76 pKa = 3.71YY77 pKa = 10.66SITLSAYY84 pKa = 9.83DD85 pKa = 3.74GKK87 pKa = 9.77LTTSVSVVITVTNDD101 pKa = 2.83VSDD104 pKa = 4.63DD105 pKa = 3.51EE106 pKa = 4.53AAEE109 pKa = 3.99RR110 pKa = 11.84TLFGLSVDD118 pKa = 3.66VGEE121 pKa = 4.95VGTPTYY127 pKa = 10.59DD128 pKa = 4.88RR129 pKa = 11.84PSTWDD134 pKa = 4.94DD135 pKa = 3.4EE136 pKa = 5.4DD137 pKa = 5.12GDD139 pKa = 4.85CISDD143 pKa = 3.21RR144 pKa = 11.84HH145 pKa = 6.31EE146 pKa = 3.9ILIAQHH152 pKa = 6.63LEE154 pKa = 4.09GEE156 pKa = 4.51GAYY159 pKa = 9.52PLVMSSNGCFVEE171 pKa = 4.25TGRR174 pKa = 11.84WLDD177 pKa = 3.77PYY179 pKa = 11.63DD180 pKa = 3.92NIYY183 pKa = 10.47YY184 pKa = 10.42YY185 pKa = 10.65SASDD189 pKa = 3.5VQIDD193 pKa = 3.87HH194 pKa = 6.89LVALYY199 pKa = 9.47EE200 pKa = 4.2SWISGLGNLDD210 pKa = 3.28AALQRR215 pKa = 11.84RR216 pKa = 11.84FANTGSLEE224 pKa = 4.07EE225 pKa = 4.33GVLPEE230 pKa = 4.09TSHH233 pKa = 6.73NFLAVGASSNGEE245 pKa = 4.19KK246 pKa = 10.62GSSDD250 pKa = 3.11PTEE253 pKa = 3.54WMPRR257 pKa = 11.84NEE259 pKa = 4.73AYY261 pKa = 10.37HH262 pKa = 6.94CIYY265 pKa = 9.96LKK267 pKa = 10.59KK268 pKa = 9.62WVLTKK273 pKa = 10.58SQNGLLIDD281 pKa = 3.49QAEE284 pKa = 3.96FDD286 pKa = 4.64FIEE289 pKa = 5.43GRR291 pKa = 11.84AADD294 pKa = 4.72CGAEE298 pKa = 4.31PLPEE302 pKa = 4.7LPPNPP307 pKa = 4.09
MM1 pKa = 7.04NRR3 pKa = 11.84APVFDD8 pKa = 3.74QASYY12 pKa = 11.1ALSVQEE18 pKa = 4.13NTGTTISGISFSDD31 pKa = 3.67PDD33 pKa = 4.34ADD35 pKa = 5.47PITLSISGTDD45 pKa = 3.33SEE47 pKa = 4.86AFSVTSIGSLAFITPPDD64 pKa = 3.91FEE66 pKa = 4.83TPADD70 pKa = 3.49SDD72 pKa = 3.8QNNEE76 pKa = 3.71YY77 pKa = 10.66SITLSAYY84 pKa = 9.83DD85 pKa = 3.74GKK87 pKa = 9.77LTTSVSVVITVTNDD101 pKa = 2.83VSDD104 pKa = 4.63DD105 pKa = 3.51EE106 pKa = 4.53AAEE109 pKa = 3.99RR110 pKa = 11.84TLFGLSVDD118 pKa = 3.66VGEE121 pKa = 4.95VGTPTYY127 pKa = 10.59DD128 pKa = 4.88RR129 pKa = 11.84PSTWDD134 pKa = 4.94DD135 pKa = 3.4EE136 pKa = 5.4DD137 pKa = 5.12GDD139 pKa = 4.85CISDD143 pKa = 3.21RR144 pKa = 11.84HH145 pKa = 6.31EE146 pKa = 3.9ILIAQHH152 pKa = 6.63LEE154 pKa = 4.09GEE156 pKa = 4.51GAYY159 pKa = 9.52PLVMSSNGCFVEE171 pKa = 4.25TGRR174 pKa = 11.84WLDD177 pKa = 3.77PYY179 pKa = 11.63DD180 pKa = 3.92NIYY183 pKa = 10.47YY184 pKa = 10.42YY185 pKa = 10.65SASDD189 pKa = 3.5VQIDD193 pKa = 3.87HH194 pKa = 6.89LVALYY199 pKa = 9.47EE200 pKa = 4.2SWISGLGNLDD210 pKa = 3.28AALQRR215 pKa = 11.84RR216 pKa = 11.84FANTGSLEE224 pKa = 4.07EE225 pKa = 4.33GVLPEE230 pKa = 4.09TSHH233 pKa = 6.73NFLAVGASSNGEE245 pKa = 4.19KK246 pKa = 10.62GSSDD250 pKa = 3.11PTEE253 pKa = 3.54WMPRR257 pKa = 11.84NEE259 pKa = 4.73AYY261 pKa = 10.37HH262 pKa = 6.94CIYY265 pKa = 9.96LKK267 pKa = 10.59KK268 pKa = 9.62WVLTKK273 pKa = 10.58SQNGLLIDD281 pKa = 3.49QAEE284 pKa = 3.96FDD286 pKa = 4.64FIEE289 pKa = 5.43GRR291 pKa = 11.84AADD294 pKa = 4.72CGAEE298 pKa = 4.31PLPEE302 pKa = 4.7LPPNPP307 pKa = 4.09
Molecular weight: 33.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H3NXZ9|H3NXZ9_9GAMM Putative glycosyltransferase OS=gamma proteobacterium HIMB55 OX=745014 GN=OMB55_00004400 PE=4 SV=1
MM1 pKa = 7.96LMRR4 pKa = 11.84EE5 pKa = 3.94RR6 pKa = 11.84FALYY10 pKa = 10.06RR11 pKa = 11.84SIGLGQIFVNCIFWLFRR28 pKa = 11.84KK29 pKa = 9.15QRR31 pKa = 11.84PRR33 pKa = 11.84FLVHH37 pKa = 5.74FTSVVIKK44 pKa = 9.89PGNITFSRR52 pKa = 11.84DD53 pKa = 2.96VTTLKK58 pKa = 10.8SLAVSAGCYY67 pKa = 8.4FQANNGIHH75 pKa = 7.01LGRR78 pKa = 11.84RR79 pKa = 11.84CLFAPGVKK87 pKa = 9.51IISSNHH93 pKa = 6.69DD94 pKa = 3.04IYY96 pKa = 11.54SADD99 pKa = 3.94RR100 pKa = 11.84KK101 pKa = 10.61SVVADD106 pKa = 5.04GITIGNDD113 pKa = 2.54VWIGANSVILPGVNIADD130 pKa = 3.66RR131 pKa = 11.84VVIGAGSVVTKK142 pKa = 10.72SISTPGAVVGGNPARR157 pKa = 11.84LLHH160 pKa = 6.28
MM1 pKa = 7.96LMRR4 pKa = 11.84EE5 pKa = 3.94RR6 pKa = 11.84FALYY10 pKa = 10.06RR11 pKa = 11.84SIGLGQIFVNCIFWLFRR28 pKa = 11.84KK29 pKa = 9.15QRR31 pKa = 11.84PRR33 pKa = 11.84FLVHH37 pKa = 5.74FTSVVIKK44 pKa = 9.89PGNITFSRR52 pKa = 11.84DD53 pKa = 2.96VTTLKK58 pKa = 10.8SLAVSAGCYY67 pKa = 8.4FQANNGIHH75 pKa = 7.01LGRR78 pKa = 11.84RR79 pKa = 11.84CLFAPGVKK87 pKa = 9.51IISSNHH93 pKa = 6.69DD94 pKa = 3.04IYY96 pKa = 11.54SADD99 pKa = 3.94RR100 pKa = 11.84KK101 pKa = 10.61SVVADD106 pKa = 5.04GITIGNDD113 pKa = 2.54VWIGANSVILPGVNIADD130 pKa = 3.66RR131 pKa = 11.84VVIGAGSVVTKK142 pKa = 10.72SISTPGAVVGGNPARR157 pKa = 11.84LLHH160 pKa = 6.28
Molecular weight: 17.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
835561 |
29 |
2208 |
338.3 |
36.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.443 ± 0.054 | 0.964 ± 0.016 |
6.151 ± 0.038 | 6.48 ± 0.037 |
3.838 ± 0.032 | 8.006 ± 0.042 |
2.026 ± 0.026 | 5.395 ± 0.036 |
3.492 ± 0.034 | 9.926 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.555 ± 0.024 | 3.348 ± 0.031 |
4.458 ± 0.032 | 3.525 ± 0.026 |
5.703 ± 0.041 | 6.752 ± 0.039 |
5.524 ± 0.032 | 7.471 ± 0.04 |
1.37 ± 0.018 | 2.574 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
