
Anopheles stephensi (Indo-Pakistan malaria mosquito)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera;
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
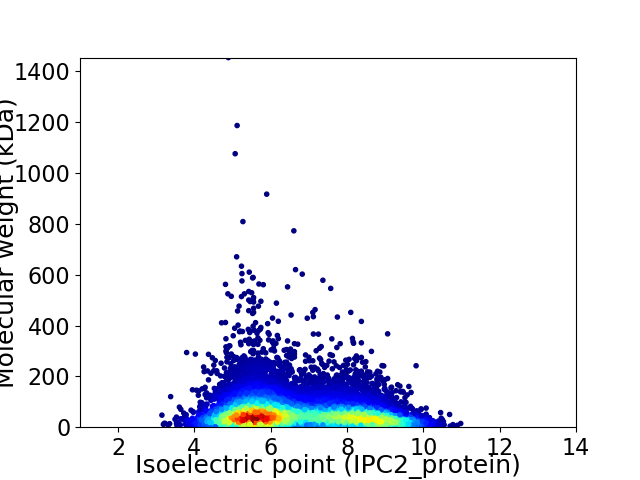
Virtual 2D-PAGE plot for 11765 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A182XYE5|A0A182XYE5_ANOST Uncharacterized protein OS=Anopheles stephensi OX=30069 PE=4 SV=1
MM1 pKa = 7.51CCAPMSDD8 pKa = 4.31YY9 pKa = 9.03CTKK12 pKa = 10.15MALLDD17 pKa = 3.87DD18 pKa = 4.44TNGNTVNINNQPSGEE33 pKa = 4.19AGPGDD38 pKa = 3.8GGGGGMAPATGDD50 pKa = 3.8SVTMLHH56 pKa = 6.54SPSNDD61 pKa = 3.33DD62 pKa = 3.76LSQSLSEE69 pKa = 4.12YY70 pKa = 10.0TDD72 pKa = 3.56ADD74 pKa = 3.78EE75 pKa = 5.73SISAPTEE82 pKa = 3.6LLAEE86 pKa = 4.23FLSAIMIRR94 pKa = 11.84DD95 pKa = 3.45YY96 pKa = 11.94VNALKK101 pKa = 10.6YY102 pKa = 10.31CKK104 pKa = 9.93QILQYY109 pKa = 10.68EE110 pKa = 4.53PNNTTARR117 pKa = 11.84DD118 pKa = 4.66FYY120 pKa = 10.89PHH122 pKa = 7.03ILSKK126 pKa = 10.41ISEE129 pKa = 4.26MQNNEE134 pKa = 3.57LSGEE138 pKa = 3.91SDD140 pKa = 2.95EE141 pKa = 5.59NYY143 pKa = 10.76NFNCSSPLASSASSPSSCSVSSSPGSSRR171 pKa = 11.84SSRR174 pKa = 11.84SGSSSSSSSGCDD186 pKa = 3.27NIDD189 pKa = 3.46EE190 pKa = 4.72VIMVCSSDD198 pKa = 3.39SDD200 pKa = 3.78EE201 pKa = 5.07SSEE204 pKa = 4.39ACCSLPDD211 pKa = 4.39SVGDD215 pKa = 3.69EE216 pKa = 4.24AKK218 pKa = 10.72GEE220 pKa = 4.08PRR222 pKa = 11.84SEE224 pKa = 4.06PVGEE228 pKa = 4.17RR229 pKa = 11.84QVVTAVGAIPATGGSRR245 pKa = 11.84SLSDD249 pKa = 3.79ANTSQSYY256 pKa = 11.1SSLLLEE262 pKa = 5.6DD263 pKa = 4.09EE264 pKa = 4.9EE265 pKa = 6.15KK266 pKa = 11.13DD267 pKa = 3.51LTLSDD272 pKa = 3.41ISNLNIEE279 pKa = 4.45EE280 pKa = 4.98DD281 pKa = 3.82EE282 pKa = 4.35TSNDD286 pKa = 3.46CGGSLEE292 pKa = 4.38NSYY295 pKa = 9.63TPLDD299 pKa = 3.76VSAPPVVKK307 pKa = 10.35PPVPTTNLASATVFASRR324 pKa = 11.84IVAMLRR330 pKa = 11.84AKK332 pKa = 9.96VIPSKK337 pKa = 10.87AAGDD341 pKa = 3.69
MM1 pKa = 7.51CCAPMSDD8 pKa = 4.31YY9 pKa = 9.03CTKK12 pKa = 10.15MALLDD17 pKa = 3.87DD18 pKa = 4.44TNGNTVNINNQPSGEE33 pKa = 4.19AGPGDD38 pKa = 3.8GGGGGMAPATGDD50 pKa = 3.8SVTMLHH56 pKa = 6.54SPSNDD61 pKa = 3.33DD62 pKa = 3.76LSQSLSEE69 pKa = 4.12YY70 pKa = 10.0TDD72 pKa = 3.56ADD74 pKa = 3.78EE75 pKa = 5.73SISAPTEE82 pKa = 3.6LLAEE86 pKa = 4.23FLSAIMIRR94 pKa = 11.84DD95 pKa = 3.45YY96 pKa = 11.94VNALKK101 pKa = 10.6YY102 pKa = 10.31CKK104 pKa = 9.93QILQYY109 pKa = 10.68EE110 pKa = 4.53PNNTTARR117 pKa = 11.84DD118 pKa = 4.66FYY120 pKa = 10.89PHH122 pKa = 7.03ILSKK126 pKa = 10.41ISEE129 pKa = 4.26MQNNEE134 pKa = 3.57LSGEE138 pKa = 3.91SDD140 pKa = 2.95EE141 pKa = 5.59NYY143 pKa = 10.76NFNCSSPLASSASSPSSCSVSSSPGSSRR171 pKa = 11.84SSRR174 pKa = 11.84SGSSSSSSSGCDD186 pKa = 3.27NIDD189 pKa = 3.46EE190 pKa = 4.72VIMVCSSDD198 pKa = 3.39SDD200 pKa = 3.78EE201 pKa = 5.07SSEE204 pKa = 4.39ACCSLPDD211 pKa = 4.39SVGDD215 pKa = 3.69EE216 pKa = 4.24AKK218 pKa = 10.72GEE220 pKa = 4.08PRR222 pKa = 11.84SEE224 pKa = 4.06PVGEE228 pKa = 4.17RR229 pKa = 11.84QVVTAVGAIPATGGSRR245 pKa = 11.84SLSDD249 pKa = 3.79ANTSQSYY256 pKa = 11.1SSLLLEE262 pKa = 5.6DD263 pKa = 4.09EE264 pKa = 4.9EE265 pKa = 6.15KK266 pKa = 11.13DD267 pKa = 3.51LTLSDD272 pKa = 3.41ISNLNIEE279 pKa = 4.45EE280 pKa = 4.98DD281 pKa = 3.82EE282 pKa = 4.35TSNDD286 pKa = 3.46CGGSLEE292 pKa = 4.38NSYY295 pKa = 9.63TPLDD299 pKa = 3.76VSAPPVVKK307 pKa = 10.35PPVPTTNLASATVFASRR324 pKa = 11.84IVAMLRR330 pKa = 11.84AKK332 pKa = 9.96VIPSKK337 pKa = 10.87AAGDD341 pKa = 3.69
Molecular weight: 35.48 kDa
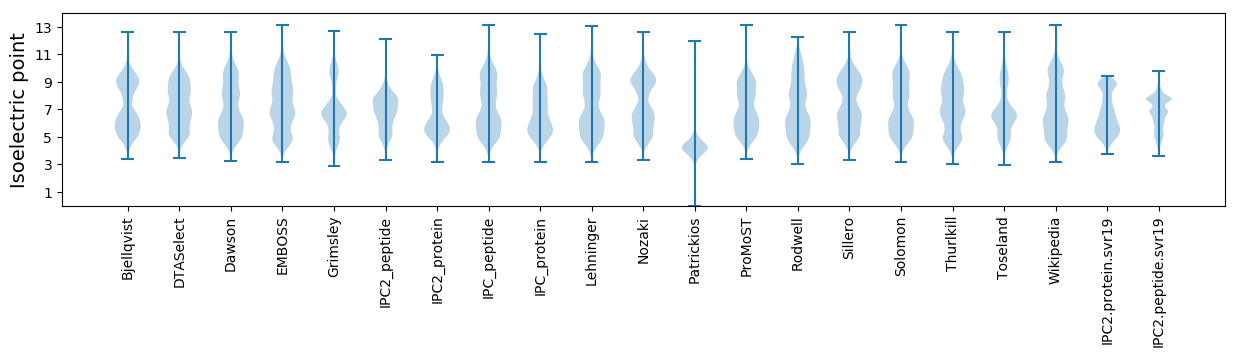
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A182XV26|A0A182XV26_ANOST 39S ribosomal protein L34 mitochondrial OS=Anopheles stephensi OX=30069 PE=3 SV=1
LL1 pKa = 7.48SRR3 pKa = 11.84LPLLPMLPRR12 pKa = 11.84LPTLTIKK19 pKa = 9.09EE20 pKa = 4.55TTSRR24 pKa = 11.84PSRR27 pKa = 11.84LSRR30 pKa = 11.84PSTLRR35 pKa = 11.84SRR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 9.48LAMWPPTVVLSMRR52 pKa = 11.84LLWPVMPSTRR62 pKa = 11.84SRR64 pKa = 3.28
LL1 pKa = 7.48SRR3 pKa = 11.84LPLLPMLPRR12 pKa = 11.84LPTLTIKK19 pKa = 9.09EE20 pKa = 4.55TTSRR24 pKa = 11.84PSRR27 pKa = 11.84LSRR30 pKa = 11.84PSTLRR35 pKa = 11.84SRR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 9.48LAMWPPTVVLSMRR52 pKa = 11.84LLWPVMPSTRR62 pKa = 11.84SRR64 pKa = 3.28
Molecular weight: 7.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6746010 |
23 |
12716 |
573.4 |
63.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.411 ± 0.021 | 1.849 ± 0.028 |
5.339 ± 0.018 | 6.253 ± 0.028 |
3.505 ± 0.02 | 6.656 ± 0.034 |
2.702 ± 0.015 | 4.876 ± 0.021 |
5.27 ± 0.024 | 8.909 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.279 ± 0.01 | 4.5 ± 0.017 |
5.452 ± 0.026 | 4.782 ± 0.032 |
5.631 ± 0.021 | 8.215 ± 0.038 |
6.085 ± 0.019 | 6.263 ± 0.02 |
0.971 ± 0.007 | 3.05 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
