
Beihai horseshoe crab virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.77
Get precalculated fractions of proteins
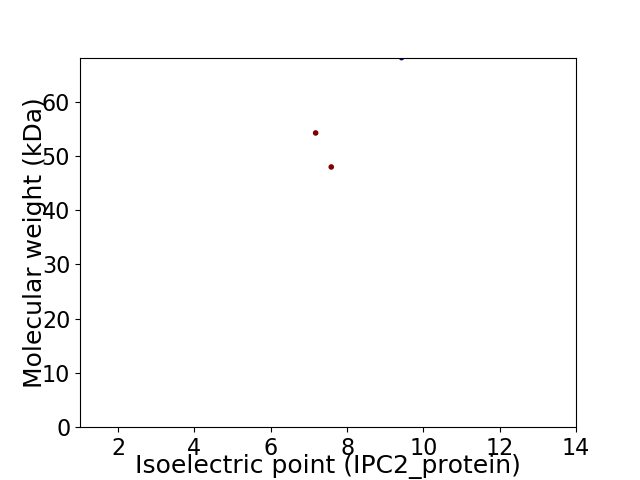
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFV0|A0A1L3KFV0_9VIRU Uncharacterized protein OS=Beihai horseshoe crab virus 1 OX=1922392 PE=4 SV=1
MM1 pKa = 7.78RR2 pKa = 11.84PSQTNTAGPEE12 pKa = 3.44WDD14 pKa = 3.89RR15 pKa = 11.84YY16 pKa = 8.27LTEE19 pKa = 4.75LGRR22 pKa = 11.84LATAIGRR29 pKa = 11.84VPKK32 pKa = 10.66ASTKK36 pKa = 10.68AILAGLKK43 pKa = 9.6GRR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 9.51RR48 pKa = 11.84RR49 pKa = 11.84YY50 pKa = 7.78HH51 pKa = 5.92QGFSTLIAKK60 pKa = 9.97GIGRR64 pKa = 11.84RR65 pKa = 11.84DD66 pKa = 3.42AYY68 pKa = 9.69IKK70 pKa = 10.88SMQKK74 pKa = 10.21LEE76 pKa = 4.19LYY78 pKa = 9.96EE79 pKa = 4.08VEE81 pKa = 4.87KK82 pKa = 11.01LDD84 pKa = 3.97SKK86 pKa = 10.56EE87 pKa = 3.76NRR89 pKa = 11.84AIQYY93 pKa = 10.33RR94 pKa = 11.84SVIYY98 pKa = 9.37NAAMARR104 pKa = 11.84EE105 pKa = 4.08LHH107 pKa = 6.38CFSKK111 pKa = 10.74RR112 pKa = 11.84LAVVHH117 pKa = 6.15SLNDD121 pKa = 3.49TNTPITTKK129 pKa = 10.7GMTPRR134 pKa = 11.84QMAEE138 pKa = 3.86KK139 pKa = 10.05IVFEE143 pKa = 4.16WSTYY147 pKa = 8.35EE148 pKa = 4.81DD149 pKa = 4.04PVALLLDD156 pKa = 4.28FSRR159 pKa = 11.84MDD161 pKa = 3.17GSYY164 pKa = 10.58TIDD167 pKa = 3.47TIRR170 pKa = 11.84AEE172 pKa = 4.05HH173 pKa = 6.54AVYY176 pKa = 10.11KK177 pKa = 9.81YY178 pKa = 10.29CRR180 pKa = 11.84PSKK183 pKa = 10.27RR184 pKa = 11.84LSWLLKK190 pKa = 8.19QQEE193 pKa = 3.92RR194 pKa = 11.84SYY196 pKa = 11.33GVTEE200 pKa = 4.23GGVKK204 pKa = 10.1YY205 pKa = 8.75VTYY208 pKa = 10.44GKK210 pKa = 10.32RR211 pKa = 11.84SSGDD215 pKa = 3.4YY216 pKa = 8.49NTSDD220 pKa = 3.28GNTITNYY227 pKa = 10.54AATSSWCGSVRR238 pKa = 11.84RR239 pKa = 11.84SFTGIGDD246 pKa = 4.05DD247 pKa = 3.27NVVIMEE253 pKa = 4.24RR254 pKa = 11.84NSYY257 pKa = 10.48YY258 pKa = 10.67SLPPVEE264 pKa = 5.77GFFLEE269 pKa = 4.24LGFVVEE275 pKa = 4.17PVIAEE280 pKa = 4.1RR281 pKa = 11.84VCDD284 pKa = 4.24IEE286 pKa = 4.41YY287 pKa = 9.94CQSKK291 pKa = 10.53IMFDD295 pKa = 3.52DD296 pKa = 3.26QGPIFVRR303 pKa = 11.84DD304 pKa = 3.3WKK306 pKa = 11.03KK307 pKa = 10.4VVEE310 pKa = 4.38TVKK313 pKa = 10.6LSPVHH318 pKa = 6.38YY319 pKa = 9.93DD320 pKa = 3.23ARR322 pKa = 11.84LCKK325 pKa = 9.74QVALANLLCQLAIYY339 pKa = 10.5NDD341 pKa = 3.44IPIIGPYY348 pKa = 9.89CYY350 pKa = 10.17RR351 pKa = 11.84NLKK354 pKa = 10.04HH355 pKa = 6.53YY356 pKa = 11.04DD357 pKa = 3.19GVTPRR362 pKa = 11.84LPEE365 pKa = 3.95YY366 pKa = 11.06LDD368 pKa = 3.99FKK370 pKa = 10.39MNYY373 pKa = 8.23IKK375 pKa = 10.66RR376 pKa = 11.84VDD378 pKa = 3.24ISKK381 pKa = 11.13LEE383 pKa = 3.81VHH385 pKa = 6.19EE386 pKa = 4.04PKK388 pKa = 10.22IRR390 pKa = 11.84DD391 pKa = 3.63TVRR394 pKa = 11.84MSFHH398 pKa = 6.2RR399 pKa = 11.84TFGVDD404 pKa = 3.07PTMQMWLEE412 pKa = 4.07EE413 pKa = 4.25SLDD416 pKa = 3.75YY417 pKa = 11.47CPVEE421 pKa = 4.64NNAKK425 pKa = 9.27QPKK428 pKa = 9.43IKK430 pKa = 9.11VTKK433 pKa = 9.89SLEE436 pKa = 4.19EE437 pKa = 3.72IVDD440 pKa = 4.04FEE442 pKa = 5.18EE443 pKa = 4.56NFTVEE448 pKa = 3.95PCDD451 pKa = 4.27CGEE454 pKa = 4.38CPDD457 pKa = 4.06YY458 pKa = 11.29SWNEE462 pKa = 3.76EE463 pKa = 4.13LLGHH467 pKa = 6.84LRR469 pKa = 11.84EE470 pKa = 4.6CC471 pKa = 5.06
MM1 pKa = 7.78RR2 pKa = 11.84PSQTNTAGPEE12 pKa = 3.44WDD14 pKa = 3.89RR15 pKa = 11.84YY16 pKa = 8.27LTEE19 pKa = 4.75LGRR22 pKa = 11.84LATAIGRR29 pKa = 11.84VPKK32 pKa = 10.66ASTKK36 pKa = 10.68AILAGLKK43 pKa = 9.6GRR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 9.51RR48 pKa = 11.84RR49 pKa = 11.84YY50 pKa = 7.78HH51 pKa = 5.92QGFSTLIAKK60 pKa = 9.97GIGRR64 pKa = 11.84RR65 pKa = 11.84DD66 pKa = 3.42AYY68 pKa = 9.69IKK70 pKa = 10.88SMQKK74 pKa = 10.21LEE76 pKa = 4.19LYY78 pKa = 9.96EE79 pKa = 4.08VEE81 pKa = 4.87KK82 pKa = 11.01LDD84 pKa = 3.97SKK86 pKa = 10.56EE87 pKa = 3.76NRR89 pKa = 11.84AIQYY93 pKa = 10.33RR94 pKa = 11.84SVIYY98 pKa = 9.37NAAMARR104 pKa = 11.84EE105 pKa = 4.08LHH107 pKa = 6.38CFSKK111 pKa = 10.74RR112 pKa = 11.84LAVVHH117 pKa = 6.15SLNDD121 pKa = 3.49TNTPITTKK129 pKa = 10.7GMTPRR134 pKa = 11.84QMAEE138 pKa = 3.86KK139 pKa = 10.05IVFEE143 pKa = 4.16WSTYY147 pKa = 8.35EE148 pKa = 4.81DD149 pKa = 4.04PVALLLDD156 pKa = 4.28FSRR159 pKa = 11.84MDD161 pKa = 3.17GSYY164 pKa = 10.58TIDD167 pKa = 3.47TIRR170 pKa = 11.84AEE172 pKa = 4.05HH173 pKa = 6.54AVYY176 pKa = 10.11KK177 pKa = 9.81YY178 pKa = 10.29CRR180 pKa = 11.84PSKK183 pKa = 10.27RR184 pKa = 11.84LSWLLKK190 pKa = 8.19QQEE193 pKa = 3.92RR194 pKa = 11.84SYY196 pKa = 11.33GVTEE200 pKa = 4.23GGVKK204 pKa = 10.1YY205 pKa = 8.75VTYY208 pKa = 10.44GKK210 pKa = 10.32RR211 pKa = 11.84SSGDD215 pKa = 3.4YY216 pKa = 8.49NTSDD220 pKa = 3.28GNTITNYY227 pKa = 10.54AATSSWCGSVRR238 pKa = 11.84RR239 pKa = 11.84SFTGIGDD246 pKa = 4.05DD247 pKa = 3.27NVVIMEE253 pKa = 4.24RR254 pKa = 11.84NSYY257 pKa = 10.48YY258 pKa = 10.67SLPPVEE264 pKa = 5.77GFFLEE269 pKa = 4.24LGFVVEE275 pKa = 4.17PVIAEE280 pKa = 4.1RR281 pKa = 11.84VCDD284 pKa = 4.24IEE286 pKa = 4.41YY287 pKa = 9.94CQSKK291 pKa = 10.53IMFDD295 pKa = 3.52DD296 pKa = 3.26QGPIFVRR303 pKa = 11.84DD304 pKa = 3.3WKK306 pKa = 11.03KK307 pKa = 10.4VVEE310 pKa = 4.38TVKK313 pKa = 10.6LSPVHH318 pKa = 6.38YY319 pKa = 9.93DD320 pKa = 3.23ARR322 pKa = 11.84LCKK325 pKa = 9.74QVALANLLCQLAIYY339 pKa = 10.5NDD341 pKa = 3.44IPIIGPYY348 pKa = 9.89CYY350 pKa = 10.17RR351 pKa = 11.84NLKK354 pKa = 10.04HH355 pKa = 6.53YY356 pKa = 11.04DD357 pKa = 3.19GVTPRR362 pKa = 11.84LPEE365 pKa = 3.95YY366 pKa = 11.06LDD368 pKa = 3.99FKK370 pKa = 10.39MNYY373 pKa = 8.23IKK375 pKa = 10.66RR376 pKa = 11.84VDD378 pKa = 3.24ISKK381 pKa = 11.13LEE383 pKa = 3.81VHH385 pKa = 6.19EE386 pKa = 4.04PKK388 pKa = 10.22IRR390 pKa = 11.84DD391 pKa = 3.63TVRR394 pKa = 11.84MSFHH398 pKa = 6.2RR399 pKa = 11.84TFGVDD404 pKa = 3.07PTMQMWLEE412 pKa = 4.07EE413 pKa = 4.25SLDD416 pKa = 3.75YY417 pKa = 11.47CPVEE421 pKa = 4.64NNAKK425 pKa = 9.27QPKK428 pKa = 9.43IKK430 pKa = 9.11VTKK433 pKa = 9.89SLEE436 pKa = 4.19EE437 pKa = 3.72IVDD440 pKa = 4.04FEE442 pKa = 5.18EE443 pKa = 4.56NFTVEE448 pKa = 3.95PCDD451 pKa = 4.27CGEE454 pKa = 4.38CPDD457 pKa = 4.06YY458 pKa = 11.29SWNEE462 pKa = 3.76EE463 pKa = 4.13LLGHH467 pKa = 6.84LRR469 pKa = 11.84EE470 pKa = 4.6CC471 pKa = 5.06
Molecular weight: 54.22 kDa
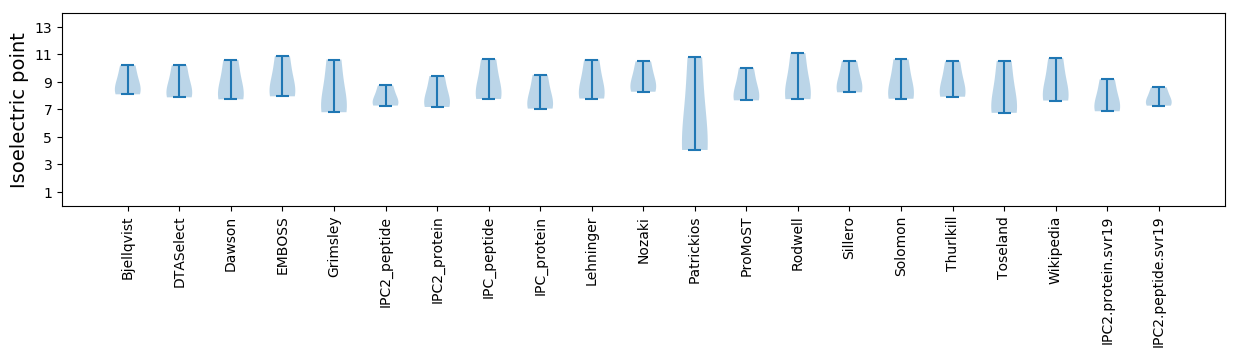
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG54|A0A1L3KG54_9VIRU Capsid protein OS=Beihai horseshoe crab virus 1 OX=1922392 PE=4 SV=1
MM1 pKa = 7.35SNKK4 pKa = 7.94TVKK7 pKa = 10.41PGVVKK12 pKa = 10.5RR13 pKa = 11.84KK14 pKa = 10.09KK15 pKa = 8.48PARR18 pKa = 11.84PNRR21 pKa = 11.84EE22 pKa = 3.65LKK24 pKa = 9.94QAPKK28 pKa = 10.38SDD30 pKa = 4.35DD31 pKa = 3.43PALPLTVVINGVEE44 pKa = 4.41VTSGPGTVISIWRR57 pKa = 11.84GGKK60 pKa = 8.57RR61 pKa = 11.84WYY63 pKa = 8.84THH65 pKa = 6.1GSPPKK70 pKa = 8.69SGSVGTVVPGVSYY83 pKa = 11.14AEE85 pKa = 4.33KK86 pKa = 10.69VSTDD90 pKa = 3.22PKK92 pKa = 10.21EE93 pKa = 3.99EE94 pKa = 4.01SRR96 pKa = 11.84KK97 pKa = 9.87KK98 pKa = 10.16DD99 pKa = 3.19TVPPVTAVKK108 pKa = 10.48VHH110 pKa = 6.76TISSADD116 pKa = 3.46EE117 pKa = 4.2KK118 pKa = 10.94PPSRR122 pKa = 11.84RR123 pKa = 11.84QKK125 pKa = 9.65RR126 pKa = 11.84LVVAEE131 pKa = 3.88QATAEE136 pKa = 4.28VAKK139 pKa = 10.94SGGSWNEE146 pKa = 3.06IFRR149 pKa = 11.84RR150 pKa = 11.84LWSARR155 pKa = 11.84SDD157 pKa = 3.3KK158 pKa = 11.15GPKK161 pKa = 9.66QKK163 pKa = 10.27MKK165 pKa = 10.66DD166 pKa = 2.84WAKK169 pKa = 10.24QVEE172 pKa = 4.52KK173 pKa = 11.01ADD175 pKa = 3.8FLAARR180 pKa = 11.84EE181 pKa = 4.11SHH183 pKa = 6.51LGKK186 pKa = 10.78ANGKK190 pKa = 4.22TLKK193 pKa = 10.13RR194 pKa = 11.84RR195 pKa = 11.84RR196 pKa = 11.84QRR198 pKa = 11.84LAAKK202 pKa = 10.32NKK204 pKa = 9.99DD205 pKa = 3.33RR206 pKa = 11.84VTAWVADD213 pKa = 3.78QNRR216 pKa = 11.84QNLTDD221 pKa = 3.81LQTIPEE227 pKa = 4.66RR228 pKa = 11.84PPTPHH233 pKa = 7.49PIILDD238 pKa = 3.75KK239 pKa = 11.31LPEE242 pKa = 4.78DD243 pKa = 4.35LDD245 pKa = 3.88EE246 pKa = 4.64VDD248 pKa = 3.57WPKK251 pKa = 11.12GEE253 pKa = 4.54PMAQQVPLPQRR264 pKa = 11.84EE265 pKa = 4.18RR266 pKa = 11.84FRR268 pKa = 11.84KK269 pKa = 10.24DD270 pKa = 2.97PLIPVGKK277 pKa = 9.96KK278 pKa = 10.28GMAKK282 pKa = 10.2LVLDD286 pKa = 3.44NAEE289 pKa = 4.15RR290 pKa = 11.84EE291 pKa = 4.04RR292 pKa = 11.84AALYY296 pKa = 10.4RR297 pKa = 11.84LARR300 pKa = 11.84EE301 pKa = 4.12QQAEE305 pKa = 4.2AEE307 pKa = 4.36KK308 pKa = 11.19ANAALAEE315 pKa = 4.62KK316 pKa = 10.05IRR318 pKa = 11.84QRR320 pKa = 11.84EE321 pKa = 3.8KK322 pKa = 9.98SEE324 pKa = 3.54EE325 pKa = 3.72RR326 pKa = 11.84RR327 pKa = 11.84FNLLHH332 pKa = 6.12KK333 pKa = 10.0RR334 pKa = 11.84EE335 pKa = 4.38TIGPPIKK342 pKa = 9.72TKK344 pKa = 10.67SKK346 pKa = 10.26EE347 pKa = 3.98LVPLGDD353 pKa = 3.76LSGLRR358 pKa = 11.84AHH360 pKa = 6.99LQAKK364 pKa = 9.14VAFKK368 pKa = 10.6PRR370 pKa = 11.84DD371 pKa = 3.42HH372 pKa = 6.88RR373 pKa = 11.84TPAWLSFEE381 pKa = 4.08AQRR384 pKa = 11.84WVADD388 pKa = 3.62HH389 pKa = 7.28DD390 pKa = 4.07TSQWDD395 pKa = 3.55PLALQTAVEE404 pKa = 4.15MMIVTCLEE412 pKa = 3.93PRR414 pKa = 11.84SVEE417 pKa = 3.9LSSEE421 pKa = 4.15KK422 pKa = 10.12TLSTRR427 pKa = 11.84SLSKK431 pKa = 10.82LNSIYY436 pKa = 10.44RR437 pKa = 11.84WGKK440 pKa = 6.15TSRR443 pKa = 11.84FEE445 pKa = 4.21SWRR448 pKa = 11.84LCADD452 pKa = 4.19FAGGFLGCRR461 pKa = 11.84VASVIFEE468 pKa = 3.75EE469 pKa = 4.08RR470 pKa = 11.84HH471 pKa = 5.66YY472 pKa = 10.74IRR474 pKa = 11.84HH475 pKa = 6.87LINSFVARR483 pKa = 11.84QCVKK487 pKa = 10.08MGWYY491 pKa = 10.1KK492 pKa = 9.41MAYY495 pKa = 9.85RR496 pKa = 11.84FDD498 pKa = 3.87KK499 pKa = 10.68PVTLGSCVRR508 pKa = 11.84DD509 pKa = 3.26NLEE512 pKa = 3.9RR513 pKa = 11.84LNPHH517 pKa = 7.47LILKK521 pKa = 9.52AKK523 pKa = 9.72KK524 pKa = 10.18ALISLTDD531 pKa = 3.58RR532 pKa = 11.84FVLALKK538 pKa = 9.79MKK540 pKa = 10.46LGYY543 pKa = 9.47QPPVPPPVRR552 pKa = 11.84LEE554 pKa = 3.96VRR556 pKa = 11.84VARR559 pKa = 11.84YY560 pKa = 9.77LMSKK564 pKa = 8.22VTLRR568 pKa = 11.84GAVLAAGFFCGTYY581 pKa = 9.75IIYY584 pKa = 10.34RR585 pKa = 11.84KK586 pKa = 9.46VGLRR590 pKa = 11.84RR591 pKa = 11.84RR592 pKa = 11.84VRR594 pKa = 11.84HH595 pKa = 6.19PEE597 pKa = 3.68QEE599 pKa = 4.46SNN601 pKa = 3.29
MM1 pKa = 7.35SNKK4 pKa = 7.94TVKK7 pKa = 10.41PGVVKK12 pKa = 10.5RR13 pKa = 11.84KK14 pKa = 10.09KK15 pKa = 8.48PARR18 pKa = 11.84PNRR21 pKa = 11.84EE22 pKa = 3.65LKK24 pKa = 9.94QAPKK28 pKa = 10.38SDD30 pKa = 4.35DD31 pKa = 3.43PALPLTVVINGVEE44 pKa = 4.41VTSGPGTVISIWRR57 pKa = 11.84GGKK60 pKa = 8.57RR61 pKa = 11.84WYY63 pKa = 8.84THH65 pKa = 6.1GSPPKK70 pKa = 8.69SGSVGTVVPGVSYY83 pKa = 11.14AEE85 pKa = 4.33KK86 pKa = 10.69VSTDD90 pKa = 3.22PKK92 pKa = 10.21EE93 pKa = 3.99EE94 pKa = 4.01SRR96 pKa = 11.84KK97 pKa = 9.87KK98 pKa = 10.16DD99 pKa = 3.19TVPPVTAVKK108 pKa = 10.48VHH110 pKa = 6.76TISSADD116 pKa = 3.46EE117 pKa = 4.2KK118 pKa = 10.94PPSRR122 pKa = 11.84RR123 pKa = 11.84QKK125 pKa = 9.65RR126 pKa = 11.84LVVAEE131 pKa = 3.88QATAEE136 pKa = 4.28VAKK139 pKa = 10.94SGGSWNEE146 pKa = 3.06IFRR149 pKa = 11.84RR150 pKa = 11.84LWSARR155 pKa = 11.84SDD157 pKa = 3.3KK158 pKa = 11.15GPKK161 pKa = 9.66QKK163 pKa = 10.27MKK165 pKa = 10.66DD166 pKa = 2.84WAKK169 pKa = 10.24QVEE172 pKa = 4.52KK173 pKa = 11.01ADD175 pKa = 3.8FLAARR180 pKa = 11.84EE181 pKa = 4.11SHH183 pKa = 6.51LGKK186 pKa = 10.78ANGKK190 pKa = 4.22TLKK193 pKa = 10.13RR194 pKa = 11.84RR195 pKa = 11.84RR196 pKa = 11.84QRR198 pKa = 11.84LAAKK202 pKa = 10.32NKK204 pKa = 9.99DD205 pKa = 3.33RR206 pKa = 11.84VTAWVADD213 pKa = 3.78QNRR216 pKa = 11.84QNLTDD221 pKa = 3.81LQTIPEE227 pKa = 4.66RR228 pKa = 11.84PPTPHH233 pKa = 7.49PIILDD238 pKa = 3.75KK239 pKa = 11.31LPEE242 pKa = 4.78DD243 pKa = 4.35LDD245 pKa = 3.88EE246 pKa = 4.64VDD248 pKa = 3.57WPKK251 pKa = 11.12GEE253 pKa = 4.54PMAQQVPLPQRR264 pKa = 11.84EE265 pKa = 4.18RR266 pKa = 11.84FRR268 pKa = 11.84KK269 pKa = 10.24DD270 pKa = 2.97PLIPVGKK277 pKa = 9.96KK278 pKa = 10.28GMAKK282 pKa = 10.2LVLDD286 pKa = 3.44NAEE289 pKa = 4.15RR290 pKa = 11.84EE291 pKa = 4.04RR292 pKa = 11.84AALYY296 pKa = 10.4RR297 pKa = 11.84LARR300 pKa = 11.84EE301 pKa = 4.12QQAEE305 pKa = 4.2AEE307 pKa = 4.36KK308 pKa = 11.19ANAALAEE315 pKa = 4.62KK316 pKa = 10.05IRR318 pKa = 11.84QRR320 pKa = 11.84EE321 pKa = 3.8KK322 pKa = 9.98SEE324 pKa = 3.54EE325 pKa = 3.72RR326 pKa = 11.84RR327 pKa = 11.84FNLLHH332 pKa = 6.12KK333 pKa = 10.0RR334 pKa = 11.84EE335 pKa = 4.38TIGPPIKK342 pKa = 9.72TKK344 pKa = 10.67SKK346 pKa = 10.26EE347 pKa = 3.98LVPLGDD353 pKa = 3.76LSGLRR358 pKa = 11.84AHH360 pKa = 6.99LQAKK364 pKa = 9.14VAFKK368 pKa = 10.6PRR370 pKa = 11.84DD371 pKa = 3.42HH372 pKa = 6.88RR373 pKa = 11.84TPAWLSFEE381 pKa = 4.08AQRR384 pKa = 11.84WVADD388 pKa = 3.62HH389 pKa = 7.28DD390 pKa = 4.07TSQWDD395 pKa = 3.55PLALQTAVEE404 pKa = 4.15MMIVTCLEE412 pKa = 3.93PRR414 pKa = 11.84SVEE417 pKa = 3.9LSSEE421 pKa = 4.15KK422 pKa = 10.12TLSTRR427 pKa = 11.84SLSKK431 pKa = 10.82LNSIYY436 pKa = 10.44RR437 pKa = 11.84WGKK440 pKa = 6.15TSRR443 pKa = 11.84FEE445 pKa = 4.21SWRR448 pKa = 11.84LCADD452 pKa = 4.19FAGGFLGCRR461 pKa = 11.84VASVIFEE468 pKa = 3.75EE469 pKa = 4.08RR470 pKa = 11.84HH471 pKa = 5.66YY472 pKa = 10.74IRR474 pKa = 11.84HH475 pKa = 6.87LINSFVARR483 pKa = 11.84QCVKK487 pKa = 10.08MGWYY491 pKa = 10.1KK492 pKa = 9.41MAYY495 pKa = 9.85RR496 pKa = 11.84FDD498 pKa = 3.87KK499 pKa = 10.68PVTLGSCVRR508 pKa = 11.84DD509 pKa = 3.26NLEE512 pKa = 3.9RR513 pKa = 11.84LNPHH517 pKa = 7.47LILKK521 pKa = 9.52AKK523 pKa = 9.72KK524 pKa = 10.18ALISLTDD531 pKa = 3.58RR532 pKa = 11.84FVLALKK538 pKa = 9.79MKK540 pKa = 10.46LGYY543 pKa = 9.47QPPVPPPVRR552 pKa = 11.84LEE554 pKa = 3.96VRR556 pKa = 11.84VARR559 pKa = 11.84YY560 pKa = 9.77LMSKK564 pKa = 8.22VTLRR568 pKa = 11.84GAVLAAGFFCGTYY581 pKa = 9.75IIYY584 pKa = 10.34RR585 pKa = 11.84KK586 pKa = 9.46VGLRR590 pKa = 11.84RR591 pKa = 11.84RR592 pKa = 11.84VRR594 pKa = 11.84HH595 pKa = 6.19PEE597 pKa = 3.68QEE599 pKa = 4.46SNN601 pKa = 3.29
Molecular weight: 68.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1523 |
451 |
601 |
507.7 |
56.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.536 ± 1.628 | 1.773 ± 0.43 |
4.662 ± 0.467 | 5.844 ± 1.07 |
2.823 ± 0.166 | 6.303 ± 0.754 |
2.232 ± 0.265 | 4.596 ± 0.541 |
7.157 ± 1.048 | 7.879 ± 0.605 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.232 ± 0.255 | 3.414 ± 0.337 |
5.975 ± 0.599 | 3.414 ± 0.279 |
7.026 ± 1.513 | 7.551 ± 1.15 |
6.172 ± 0.82 | 7.223 ± 0.422 |
1.97 ± 0.205 | 3.217 ± 1.046 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
