
Pimoid spider associated circular virus 2
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.45
Get precalculated fractions of proteins
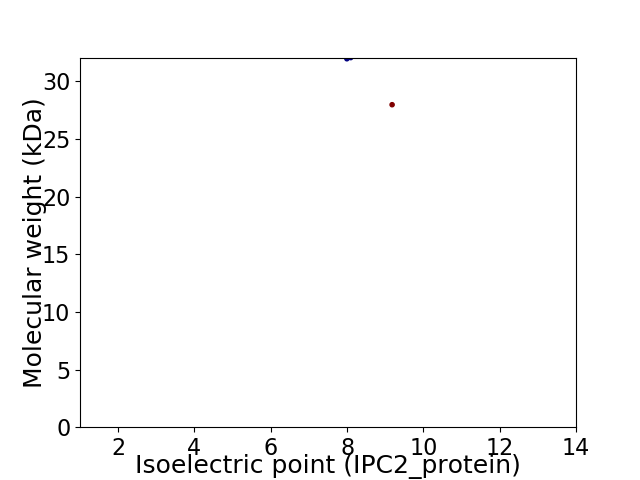
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
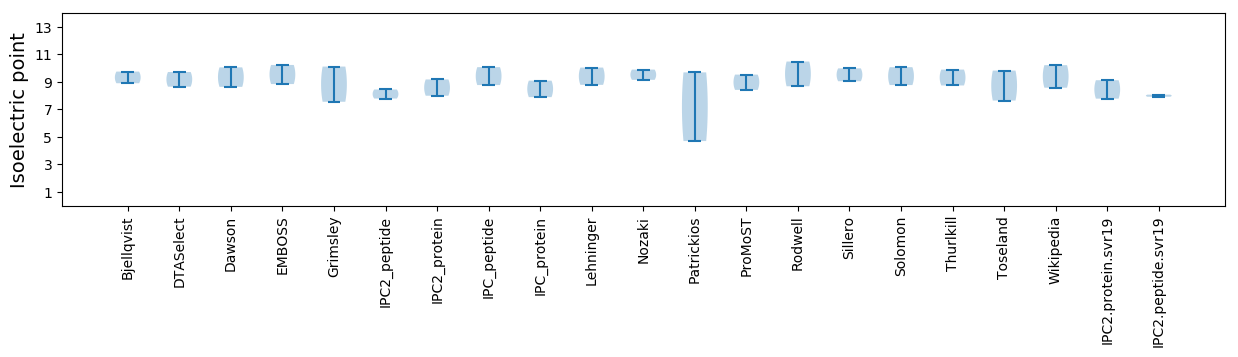
Protein with the lowest isoelectric point:
>tr|A0A346BPD3|A0A346BPD3_9VIRU Putative capsid protein OS=Pimoid spider associated circular virus 2 OX=2293298 PE=4 SV=1
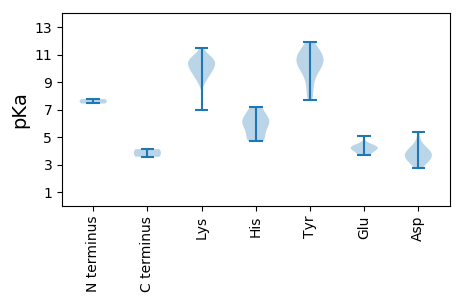
MM1 pKa = 7.44SCKK4 pKa = 9.85NWCFTWNNPDD14 pKa = 3.53LGYY17 pKa = 10.67EE18 pKa = 4.1ALLEE22 pKa = 4.12LLSSKK27 pKa = 10.58SSYY30 pKa = 10.89CIFQKK35 pKa = 10.71EE36 pKa = 3.77QGANGTVHH44 pKa = 6.56FQGFVQFTSKK54 pKa = 10.19QRR56 pKa = 11.84LSSLKK61 pKa = 10.24KK62 pKa = 9.91VLQSAHH68 pKa = 4.86WEE70 pKa = 4.28RR71 pKa = 11.84TKK73 pKa = 10.59GTAEE77 pKa = 4.05EE78 pKa = 4.02NRR80 pKa = 11.84DD81 pKa = 3.7YY82 pKa = 11.13CSKK85 pKa = 11.03ADD87 pKa = 3.81SRR89 pKa = 11.84VDD91 pKa = 3.89GPWSSGRR98 pKa = 11.84IVVKK102 pKa = 9.11GQRR105 pKa = 11.84RR106 pKa = 11.84DD107 pKa = 3.16IEE109 pKa = 4.19EE110 pKa = 3.85FRR112 pKa = 11.84DD113 pKa = 3.64AVLEE117 pKa = 4.47GSCNLDD123 pKa = 2.78LVMSFPGQVARR134 pKa = 11.84FPKK137 pKa = 10.57FIDD140 pKa = 4.01FVRR143 pKa = 11.84SAQSVRR149 pKa = 11.84RR150 pKa = 11.84VEE152 pKa = 4.3KK153 pKa = 10.32PVVEE157 pKa = 4.4CFYY160 pKa = 11.18GPSGSGKK167 pKa = 6.93TRR169 pKa = 11.84RR170 pKa = 11.84AMAVSGFDD178 pKa = 3.11STFVVSRR185 pKa = 11.84PDD187 pKa = 2.91SGRR190 pKa = 11.84PLWWDD195 pKa = 3.18GFDD198 pKa = 3.59PRR200 pKa = 11.84IHH202 pKa = 5.88ITVVLDD208 pKa = 5.37DD209 pKa = 4.31FYY211 pKa = 11.95GWIPWSFLLQLIDD224 pKa = 4.21RR225 pKa = 11.84YY226 pKa = 10.11PFQVEE231 pKa = 4.12KK232 pKa = 10.77KK233 pKa = 9.47GGKK236 pKa = 8.97IEE238 pKa = 4.4FNCKK242 pKa = 9.54NVIFTSNQHH251 pKa = 5.17PSKK254 pKa = 9.96WYY256 pKa = 10.24KK257 pKa = 10.46SVPNDD262 pKa = 3.96DD263 pKa = 3.93LTPLLRR269 pKa = 11.84RR270 pKa = 11.84IDD272 pKa = 4.14NIVLMEE278 pKa = 4.12
MM1 pKa = 7.44SCKK4 pKa = 9.85NWCFTWNNPDD14 pKa = 3.53LGYY17 pKa = 10.67EE18 pKa = 4.1ALLEE22 pKa = 4.12LLSSKK27 pKa = 10.58SSYY30 pKa = 10.89CIFQKK35 pKa = 10.71EE36 pKa = 3.77QGANGTVHH44 pKa = 6.56FQGFVQFTSKK54 pKa = 10.19QRR56 pKa = 11.84LSSLKK61 pKa = 10.24KK62 pKa = 9.91VLQSAHH68 pKa = 4.86WEE70 pKa = 4.28RR71 pKa = 11.84TKK73 pKa = 10.59GTAEE77 pKa = 4.05EE78 pKa = 4.02NRR80 pKa = 11.84DD81 pKa = 3.7YY82 pKa = 11.13CSKK85 pKa = 11.03ADD87 pKa = 3.81SRR89 pKa = 11.84VDD91 pKa = 3.89GPWSSGRR98 pKa = 11.84IVVKK102 pKa = 9.11GQRR105 pKa = 11.84RR106 pKa = 11.84DD107 pKa = 3.16IEE109 pKa = 4.19EE110 pKa = 3.85FRR112 pKa = 11.84DD113 pKa = 3.64AVLEE117 pKa = 4.47GSCNLDD123 pKa = 2.78LVMSFPGQVARR134 pKa = 11.84FPKK137 pKa = 10.57FIDD140 pKa = 4.01FVRR143 pKa = 11.84SAQSVRR149 pKa = 11.84RR150 pKa = 11.84VEE152 pKa = 4.3KK153 pKa = 10.32PVVEE157 pKa = 4.4CFYY160 pKa = 11.18GPSGSGKK167 pKa = 6.93TRR169 pKa = 11.84RR170 pKa = 11.84AMAVSGFDD178 pKa = 3.11STFVVSRR185 pKa = 11.84PDD187 pKa = 2.91SGRR190 pKa = 11.84PLWWDD195 pKa = 3.18GFDD198 pKa = 3.59PRR200 pKa = 11.84IHH202 pKa = 5.88ITVVLDD208 pKa = 5.37DD209 pKa = 4.31FYY211 pKa = 11.95GWIPWSFLLQLIDD224 pKa = 4.21RR225 pKa = 11.84YY226 pKa = 10.11PFQVEE231 pKa = 4.12KK232 pKa = 10.77KK233 pKa = 9.47GGKK236 pKa = 8.97IEE238 pKa = 4.4FNCKK242 pKa = 9.54NVIFTSNQHH251 pKa = 5.17PSKK254 pKa = 9.96WYY256 pKa = 10.24KK257 pKa = 10.46SVPNDD262 pKa = 3.96DD263 pKa = 3.93LTPLLRR269 pKa = 11.84RR270 pKa = 11.84IDD272 pKa = 4.14NIVLMEE278 pKa = 4.12
Molecular weight: 31.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPD3|A0A346BPD3_9VIRU Putative capsid protein OS=Pimoid spider associated circular virus 2 OX=2293298 PE=4 SV=1
MM1 pKa = 7.76VYY3 pKa = 9.0GTLAAVAGNYY13 pKa = 7.72AAKK16 pKa = 8.96RR17 pKa = 11.84TYY19 pKa = 11.22DD20 pKa = 3.45EE21 pKa = 3.78MHH23 pKa = 5.75NRR25 pKa = 11.84IWKK28 pKa = 9.95HH29 pKa = 4.84SGHH32 pKa = 6.26YY33 pKa = 10.96GNDD36 pKa = 3.24EE37 pKa = 4.23RR38 pKa = 11.84DD39 pKa = 3.75TNAGPSVGGYY49 pKa = 10.19GGGHH53 pKa = 6.18IKK55 pKa = 10.58SVFRR59 pKa = 11.84AKK61 pKa = 10.22KK62 pKa = 9.48HH63 pKa = 4.71SKK65 pKa = 10.21KK66 pKa = 9.74RR67 pKa = 11.84DD68 pKa = 3.52KK69 pKa = 11.47SCGTLSAASLPAIRR83 pKa = 11.84TSSGGICKK91 pKa = 9.88KK92 pKa = 10.94VYY94 pKa = 10.66DD95 pKa = 4.45FLVNTNGPIWNQSTGFTTAGPTATITSLGFSPMSVPFMGTYY136 pKa = 8.31WKK138 pKa = 10.45CLNAVEE144 pKa = 5.07HH145 pKa = 6.28VLDD148 pKa = 4.72PKK150 pKa = 9.54PTAGSATSVGQVMSFEE166 pKa = 4.54KK167 pKa = 10.55KK168 pKa = 9.34FPHH171 pKa = 7.0KK172 pKa = 10.68GRR174 pKa = 11.84IHH176 pKa = 7.17LEE178 pKa = 3.66QLRR181 pKa = 11.84GNVCVAGLSRR191 pKa = 11.84CYY193 pKa = 10.31LIKK196 pKa = 10.27EE197 pKa = 4.3APLGIDD203 pKa = 3.39PSKK206 pKa = 10.87AGPLGTIMPAFGEE219 pKa = 4.33SSGIATASKK228 pKa = 9.74IHH230 pKa = 5.88GVTYY234 pKa = 9.46TYY236 pKa = 10.0TKK238 pKa = 10.58RR239 pKa = 11.84FMMKK243 pKa = 10.07PEE245 pKa = 4.06PTSTLMARR253 pKa = 11.84PRR255 pKa = 11.84QQEE258 pKa = 3.88NAA260 pKa = 3.56
MM1 pKa = 7.76VYY3 pKa = 9.0GTLAAVAGNYY13 pKa = 7.72AAKK16 pKa = 8.96RR17 pKa = 11.84TYY19 pKa = 11.22DD20 pKa = 3.45EE21 pKa = 3.78MHH23 pKa = 5.75NRR25 pKa = 11.84IWKK28 pKa = 9.95HH29 pKa = 4.84SGHH32 pKa = 6.26YY33 pKa = 10.96GNDD36 pKa = 3.24EE37 pKa = 4.23RR38 pKa = 11.84DD39 pKa = 3.75TNAGPSVGGYY49 pKa = 10.19GGGHH53 pKa = 6.18IKK55 pKa = 10.58SVFRR59 pKa = 11.84AKK61 pKa = 10.22KK62 pKa = 9.48HH63 pKa = 4.71SKK65 pKa = 10.21KK66 pKa = 9.74RR67 pKa = 11.84DD68 pKa = 3.52KK69 pKa = 11.47SCGTLSAASLPAIRR83 pKa = 11.84TSSGGICKK91 pKa = 9.88KK92 pKa = 10.94VYY94 pKa = 10.66DD95 pKa = 4.45FLVNTNGPIWNQSTGFTTAGPTATITSLGFSPMSVPFMGTYY136 pKa = 8.31WKK138 pKa = 10.45CLNAVEE144 pKa = 5.07HH145 pKa = 6.28VLDD148 pKa = 4.72PKK150 pKa = 9.54PTAGSATSVGQVMSFEE166 pKa = 4.54KK167 pKa = 10.55KK168 pKa = 9.34FPHH171 pKa = 7.0KK172 pKa = 10.68GRR174 pKa = 11.84IHH176 pKa = 7.17LEE178 pKa = 3.66QLRR181 pKa = 11.84GNVCVAGLSRR191 pKa = 11.84CYY193 pKa = 10.31LIKK196 pKa = 10.27EE197 pKa = 4.3APLGIDD203 pKa = 3.39PSKK206 pKa = 10.87AGPLGTIMPAFGEE219 pKa = 4.33SSGIATASKK228 pKa = 9.74IHH230 pKa = 5.88GVTYY234 pKa = 9.46TYY236 pKa = 10.0TKK238 pKa = 10.58RR239 pKa = 11.84FMMKK243 pKa = 10.07PEE245 pKa = 4.06PTSTLMARR253 pKa = 11.84PRR255 pKa = 11.84QQEE258 pKa = 3.88NAA260 pKa = 3.56
Molecular weight: 27.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
538 |
260 |
278 |
269.0 |
29.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.134 ± 1.836 | 2.23 ± 0.208 |
4.647 ± 1.323 | 4.275 ± 0.551 |
5.204 ± 1.179 | 8.922 ± 1.51 |
2.416 ± 0.707 | 4.461 ± 0.104 |
7.063 ± 0.426 | 6.32 ± 0.633 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.416 ± 0.707 | 3.903 ± 0.039 |
5.576 ± 0.391 | 3.16 ± 0.837 |
5.948 ± 0.902 | 9.108 ± 0.437 |
5.948 ± 1.701 | 6.877 ± 1.01 |
2.23 ± 0.729 | 3.16 ± 0.464 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
