
Firmicutes bacterium CAG:129
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
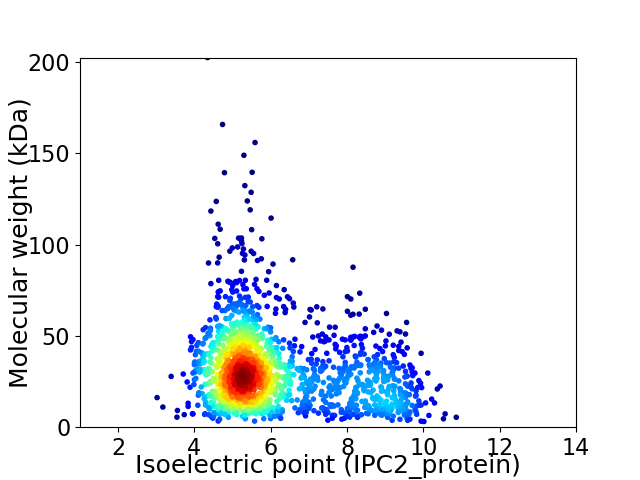
Virtual 2D-PAGE plot for 1635 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
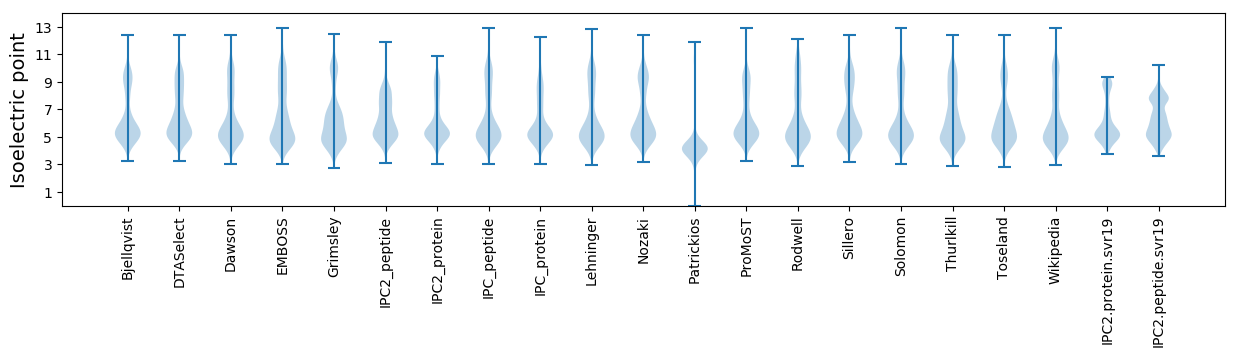
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5RXY5|R5RXY5_9FIRM CRM domain-containing protein OS=Firmicutes bacterium CAG:129 OX=1263003 GN=BN483_01176 PE=4 SV=1
MM1 pKa = 7.53KK2 pKa = 10.51KK3 pKa = 10.24FFALLLSLAMVLALVACGQKK23 pKa = 10.67DD24 pKa = 3.99PTPDD28 pKa = 3.51TPDD31 pKa = 3.68TPDD34 pKa = 3.78DD35 pKa = 4.07PQTKK39 pKa = 9.54VEE41 pKa = 4.48YY42 pKa = 9.08KK43 pKa = 10.02VAMITDD49 pKa = 3.77YY50 pKa = 11.88GDD52 pKa = 3.15ITDD55 pKa = 3.56QSFNQTTYY63 pKa = 9.78EE64 pKa = 3.85ACKK67 pKa = 10.57AFAEE71 pKa = 4.56DD72 pKa = 3.5NSIEE76 pKa = 3.92FSYY79 pKa = 10.24FKK81 pKa = 10.48PAGDD85 pKa = 3.56NTADD89 pKa = 3.34RR90 pKa = 11.84VAMIEE95 pKa = 4.04KK96 pKa = 10.43AVDD99 pKa = 2.89EE100 pKa = 4.58GFNVIVMPGYY110 pKa = 10.78AFGGAIVEE118 pKa = 4.4AAPEE122 pKa = 4.04FPDD125 pKa = 3.52VKK127 pKa = 10.82FIALDD132 pKa = 3.55VAAGDD137 pKa = 4.03LLEE140 pKa = 4.33TAVAKK145 pKa = 10.76AGEE148 pKa = 4.49TYY150 pKa = 10.49DD151 pKa = 3.78YY152 pKa = 11.19TPEE155 pKa = 3.65NWDD158 pKa = 3.39LNKK161 pKa = 10.52YY162 pKa = 10.3VDD164 pKa = 3.84MSNVYY169 pKa = 9.5CAVYY173 pKa = 10.27QEE175 pKa = 4.43EE176 pKa = 4.89LCGYY180 pKa = 7.1MAGYY184 pKa = 10.11AAVKK188 pKa = 10.3LGYY191 pKa = 10.22KK192 pKa = 10.24NLGFLGGMSVPAVVRR207 pKa = 11.84YY208 pKa = 9.95GYY210 pKa = 11.28GFVQGVDD217 pKa = 3.08AAAADD222 pKa = 4.35LGLTDD227 pKa = 3.7VKK229 pKa = 11.23VNYY232 pKa = 9.77IYY234 pKa = 10.94GGQFFGDD241 pKa = 3.47ADD243 pKa = 3.44ITAVMDD249 pKa = 3.26TWYY252 pKa = 10.71QGGTEE257 pKa = 4.26VVFACGGGIYY267 pKa = 10.05TSAVDD272 pKa = 3.29AAKK275 pKa = 10.44KK276 pKa = 10.37VGAKK280 pKa = 10.24VIGVDD285 pKa = 3.23VDD287 pKa = 3.55QAGVIAKK294 pKa = 9.65YY295 pKa = 10.54AGVDD299 pKa = 3.38GMTVTSAMKK308 pKa = 10.1GLYY311 pKa = 8.55PATYY315 pKa = 7.84DD316 pKa = 3.29TLTDD320 pKa = 3.88VIVNGNWDD328 pKa = 3.63KK329 pKa = 11.49YY330 pKa = 8.69VGKK333 pKa = 10.12IATLGLVSGTDD344 pKa = 3.29PEE346 pKa = 4.38ANYY349 pKa = 10.28VQIPMGDD356 pKa = 3.39GTQWSDD362 pKa = 3.87SFTQDD367 pKa = 3.53DD368 pKa = 4.39YY369 pKa = 12.11KK370 pKa = 11.99AMVKK374 pKa = 10.73DD375 pKa = 3.64MFDD378 pKa = 3.36GKK380 pKa = 9.43ITVSNNTSSDD390 pKa = 3.01VSAADD395 pKa = 3.47FATVITVDD403 pKa = 3.66DD404 pKa = 3.67QGSIKK409 pKa = 10.7GG410 pKa = 3.59
MM1 pKa = 7.53KK2 pKa = 10.51KK3 pKa = 10.24FFALLLSLAMVLALVACGQKK23 pKa = 10.67DD24 pKa = 3.99PTPDD28 pKa = 3.51TPDD31 pKa = 3.68TPDD34 pKa = 3.78DD35 pKa = 4.07PQTKK39 pKa = 9.54VEE41 pKa = 4.48YY42 pKa = 9.08KK43 pKa = 10.02VAMITDD49 pKa = 3.77YY50 pKa = 11.88GDD52 pKa = 3.15ITDD55 pKa = 3.56QSFNQTTYY63 pKa = 9.78EE64 pKa = 3.85ACKK67 pKa = 10.57AFAEE71 pKa = 4.56DD72 pKa = 3.5NSIEE76 pKa = 3.92FSYY79 pKa = 10.24FKK81 pKa = 10.48PAGDD85 pKa = 3.56NTADD89 pKa = 3.34RR90 pKa = 11.84VAMIEE95 pKa = 4.04KK96 pKa = 10.43AVDD99 pKa = 2.89EE100 pKa = 4.58GFNVIVMPGYY110 pKa = 10.78AFGGAIVEE118 pKa = 4.4AAPEE122 pKa = 4.04FPDD125 pKa = 3.52VKK127 pKa = 10.82FIALDD132 pKa = 3.55VAAGDD137 pKa = 4.03LLEE140 pKa = 4.33TAVAKK145 pKa = 10.76AGEE148 pKa = 4.49TYY150 pKa = 10.49DD151 pKa = 3.78YY152 pKa = 11.19TPEE155 pKa = 3.65NWDD158 pKa = 3.39LNKK161 pKa = 10.52YY162 pKa = 10.3VDD164 pKa = 3.84MSNVYY169 pKa = 9.5CAVYY173 pKa = 10.27QEE175 pKa = 4.43EE176 pKa = 4.89LCGYY180 pKa = 7.1MAGYY184 pKa = 10.11AAVKK188 pKa = 10.3LGYY191 pKa = 10.22KK192 pKa = 10.24NLGFLGGMSVPAVVRR207 pKa = 11.84YY208 pKa = 9.95GYY210 pKa = 11.28GFVQGVDD217 pKa = 3.08AAAADD222 pKa = 4.35LGLTDD227 pKa = 3.7VKK229 pKa = 11.23VNYY232 pKa = 9.77IYY234 pKa = 10.94GGQFFGDD241 pKa = 3.47ADD243 pKa = 3.44ITAVMDD249 pKa = 3.26TWYY252 pKa = 10.71QGGTEE257 pKa = 4.26VVFACGGGIYY267 pKa = 10.05TSAVDD272 pKa = 3.29AAKK275 pKa = 10.44KK276 pKa = 10.37VGAKK280 pKa = 10.24VIGVDD285 pKa = 3.23VDD287 pKa = 3.55QAGVIAKK294 pKa = 9.65YY295 pKa = 10.54AGVDD299 pKa = 3.38GMTVTSAMKK308 pKa = 10.1GLYY311 pKa = 8.55PATYY315 pKa = 7.84DD316 pKa = 3.29TLTDD320 pKa = 3.88VIVNGNWDD328 pKa = 3.63KK329 pKa = 11.49YY330 pKa = 8.69VGKK333 pKa = 10.12IATLGLVSGTDD344 pKa = 3.29PEE346 pKa = 4.38ANYY349 pKa = 10.28VQIPMGDD356 pKa = 3.39GTQWSDD362 pKa = 3.87SFTQDD367 pKa = 3.53DD368 pKa = 4.39YY369 pKa = 12.11KK370 pKa = 11.99AMVKK374 pKa = 10.73DD375 pKa = 3.64MFDD378 pKa = 3.36GKK380 pKa = 9.43ITVSNNTSSDD390 pKa = 3.01VSAADD395 pKa = 3.47FATVITVDD403 pKa = 3.66DD404 pKa = 3.67QGSIKK409 pKa = 10.7GG410 pKa = 3.59
Molecular weight: 43.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5RW71|R5RW71_9FIRM Uncharacterized protein OS=Firmicutes bacterium CAG:129 OX=1263003 GN=BN483_00686 PE=4 SV=1
MM1 pKa = 7.31AAAVPALVLFAATTLLAVFFLAASYY26 pKa = 7.64PTLCAAAKK34 pKa = 10.05RR35 pKa = 11.84RR36 pKa = 11.84LSPRR40 pKa = 11.84AQQRR44 pKa = 11.84LRR46 pKa = 11.84FWRR49 pKa = 11.84DD50 pKa = 2.7GVTHH54 pKa = 7.06SLTRR58 pKa = 11.84WLRR61 pKa = 11.84AQAILCSLTFCQLLAGFWLLGEE83 pKa = 4.71RR84 pKa = 11.84YY85 pKa = 9.62SLLAAFLITLVDD97 pKa = 3.96ALPVFGTGTVLIPWALAEE115 pKa = 4.42LLLGSVPRR123 pKa = 11.84GAALVILYY131 pKa = 9.88LCTLTVRR138 pKa = 11.84SITEE142 pKa = 3.93PKK144 pKa = 10.19LVAAQAGLPPIASLMAMYY162 pKa = 10.53LGARR166 pKa = 11.84AFGVAGMVLFPLLLLLAAQMLRR188 pKa = 11.84TKK190 pKa = 10.71KK191 pKa = 9.99EE192 pKa = 3.57GSMM195 pKa = 3.99
MM1 pKa = 7.31AAAVPALVLFAATTLLAVFFLAASYY26 pKa = 7.64PTLCAAAKK34 pKa = 10.05RR35 pKa = 11.84RR36 pKa = 11.84LSPRR40 pKa = 11.84AQQRR44 pKa = 11.84LRR46 pKa = 11.84FWRR49 pKa = 11.84DD50 pKa = 2.7GVTHH54 pKa = 7.06SLTRR58 pKa = 11.84WLRR61 pKa = 11.84AQAILCSLTFCQLLAGFWLLGEE83 pKa = 4.71RR84 pKa = 11.84YY85 pKa = 9.62SLLAAFLITLVDD97 pKa = 3.96ALPVFGTGTVLIPWALAEE115 pKa = 4.42LLLGSVPRR123 pKa = 11.84GAALVILYY131 pKa = 9.88LCTLTVRR138 pKa = 11.84SITEE142 pKa = 3.93PKK144 pKa = 10.19LVAAQAGLPPIASLMAMYY162 pKa = 10.53LGARR166 pKa = 11.84AFGVAGMVLFPLLLLLAAQMLRR188 pKa = 11.84TKK190 pKa = 10.71KK191 pKa = 9.99EE192 pKa = 3.57GSMM195 pKa = 3.99
Molecular weight: 20.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
484243 |
29 |
1862 |
296.2 |
32.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.435 ± 0.08 | 1.697 ± 0.031 |
5.622 ± 0.053 | 6.968 ± 0.059 |
3.733 ± 0.046 | 7.958 ± 0.051 |
1.909 ± 0.037 | 5.372 ± 0.058 |
4.683 ± 0.061 | 10.174 ± 0.083 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.81 ± 0.03 | 3.094 ± 0.031 |
3.913 ± 0.039 | 2.878 ± 0.033 |
6.043 ± 0.061 | 5.272 ± 0.048 |
5.642 ± 0.061 | 7.415 ± 0.051 |
0.937 ± 0.024 | 3.436 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
