
Flavobacterium sp. HMWF030
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; unclassified Flavobacterium
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
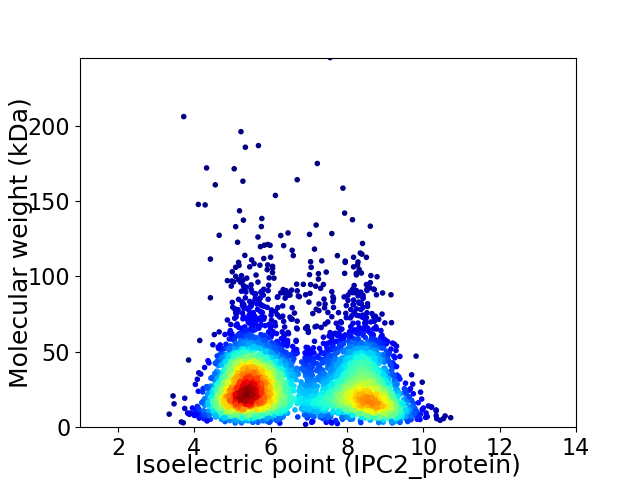
Virtual 2D-PAGE plot for 4740 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R7JQY6|A0A2R7JQY6_9FLAO Cytochrome C oxidase Cbb3 (Fragment) OS=Flavobacterium sp. HMWF030 OX=2056864 GN=DBR27_24105 PE=4 SV=1
WW1 pKa = 5.48TTGTVNNGEE10 pKa = 4.25TQTLDD15 pKa = 2.54IYY17 pKa = 11.59AKK19 pKa = 10.6VNTPSGTANEE29 pKa = 4.33YY30 pKa = 9.93TNEE33 pKa = 4.09TEE35 pKa = 4.24IVASNMPDD43 pKa = 3.52PDD45 pKa = 3.7STPNNGIKK53 pKa = 9.56TEE55 pKa = 4.31DD56 pKa = 4.51DD57 pKa = 3.49YY58 pKa = 12.2DD59 pKa = 4.62SIQINVPIADD69 pKa = 4.22LSLDD73 pKa = 3.43KK74 pKa = 10.79TVSNKK79 pKa = 9.72NPNIDD84 pKa = 3.55EE85 pKa = 4.79VITFTIQLNNAGPSQATGVAIEE107 pKa = 4.94DD108 pKa = 4.62IIPLGYY114 pKa = 10.54RR115 pKa = 11.84NITNITNGGLFSKK128 pKa = 10.83NAIKK132 pKa = 9.68WVNLTVPVGGITLTYY147 pKa = 9.66QVTVVSPHH155 pKa = 6.22GLDD158 pKa = 3.55GYY160 pKa = 11.13DD161 pKa = 3.74YY162 pKa = 11.62LNIAQVTASDD172 pKa = 4.32QYY174 pKa = 12.08DD175 pKa = 3.9PDD177 pKa = 4.12SKK179 pKa = 11.15PNNDD183 pKa = 4.34NGDD186 pKa = 3.43QSEE189 pKa = 4.41DD190 pKa = 3.91DD191 pKa = 4.11EE192 pKa = 5.91DD193 pKa = 4.63SEE195 pKa = 5.61FINISATDD203 pKa = 3.12IGINKK208 pKa = 9.4EE209 pKa = 3.46VDD211 pKa = 3.6QINVPMYY218 pKa = 9.41STVTFTITAEE228 pKa = 4.02NLGNLTATNVEE239 pKa = 4.31VLDD242 pKa = 4.44ILPKK246 pKa = 10.5GYY248 pKa = 10.17QLDD251 pKa = 4.11TFTVSAGVYY260 pKa = 10.41NSTTGIWTIPTIASKK275 pKa = 10.86GSQTLAISAKK285 pKa = 9.9VVDD288 pKa = 4.93FNDD291 pKa = 3.73YY292 pKa = 11.26LNTAQFVKK300 pKa = 9.94MDD302 pKa = 4.11QIDD305 pKa = 4.17SNSSNNQDD313 pKa = 3.38SATVAPNCLKK323 pKa = 10.36IFNEE327 pKa = 4.76FSPNDD332 pKa = 3.76DD333 pKa = 3.65GQNDD337 pKa = 3.99TFYY340 pKa = 10.59IDD342 pKa = 5.71CISQYY347 pKa = 10.69PDD349 pKa = 2.91NQLQIFNRR357 pKa = 11.84WGNLVYY363 pKa = 10.65YY364 pKa = 10.42KK365 pKa = 10.36KK366 pKa = 10.72GYY368 pKa = 9.65NNTWDD373 pKa = 3.54GKK375 pKa = 10.9ADD377 pKa = 3.75GSAKK381 pKa = 9.56TLPEE385 pKa = 3.5GTYY388 pKa = 10.11FYY390 pKa = 10.74ILDD393 pKa = 4.72LGNGSPKK400 pKa = 9.9TSGWLYY406 pKa = 11.17LKK408 pKa = 10.73
WW1 pKa = 5.48TTGTVNNGEE10 pKa = 4.25TQTLDD15 pKa = 2.54IYY17 pKa = 11.59AKK19 pKa = 10.6VNTPSGTANEE29 pKa = 4.33YY30 pKa = 9.93TNEE33 pKa = 4.09TEE35 pKa = 4.24IVASNMPDD43 pKa = 3.52PDD45 pKa = 3.7STPNNGIKK53 pKa = 9.56TEE55 pKa = 4.31DD56 pKa = 4.51DD57 pKa = 3.49YY58 pKa = 12.2DD59 pKa = 4.62SIQINVPIADD69 pKa = 4.22LSLDD73 pKa = 3.43KK74 pKa = 10.79TVSNKK79 pKa = 9.72NPNIDD84 pKa = 3.55EE85 pKa = 4.79VITFTIQLNNAGPSQATGVAIEE107 pKa = 4.94DD108 pKa = 4.62IIPLGYY114 pKa = 10.54RR115 pKa = 11.84NITNITNGGLFSKK128 pKa = 10.83NAIKK132 pKa = 9.68WVNLTVPVGGITLTYY147 pKa = 9.66QVTVVSPHH155 pKa = 6.22GLDD158 pKa = 3.55GYY160 pKa = 11.13DD161 pKa = 3.74YY162 pKa = 11.62LNIAQVTASDD172 pKa = 4.32QYY174 pKa = 12.08DD175 pKa = 3.9PDD177 pKa = 4.12SKK179 pKa = 11.15PNNDD183 pKa = 4.34NGDD186 pKa = 3.43QSEE189 pKa = 4.41DD190 pKa = 3.91DD191 pKa = 4.11EE192 pKa = 5.91DD193 pKa = 4.63SEE195 pKa = 5.61FINISATDD203 pKa = 3.12IGINKK208 pKa = 9.4EE209 pKa = 3.46VDD211 pKa = 3.6QINVPMYY218 pKa = 9.41STVTFTITAEE228 pKa = 4.02NLGNLTATNVEE239 pKa = 4.31VLDD242 pKa = 4.44ILPKK246 pKa = 10.5GYY248 pKa = 10.17QLDD251 pKa = 4.11TFTVSAGVYY260 pKa = 10.41NSTTGIWTIPTIASKK275 pKa = 10.86GSQTLAISAKK285 pKa = 9.9VVDD288 pKa = 4.93FNDD291 pKa = 3.73YY292 pKa = 11.26LNTAQFVKK300 pKa = 9.94MDD302 pKa = 4.11QIDD305 pKa = 4.17SNSSNNQDD313 pKa = 3.38SATVAPNCLKK323 pKa = 10.36IFNEE327 pKa = 4.76FSPNDD332 pKa = 3.76DD333 pKa = 3.65GQNDD337 pKa = 3.99TFYY340 pKa = 10.59IDD342 pKa = 5.71CISQYY347 pKa = 10.69PDD349 pKa = 2.91NQLQIFNRR357 pKa = 11.84WGNLVYY363 pKa = 10.65YY364 pKa = 10.42KK365 pKa = 10.36KK366 pKa = 10.72GYY368 pKa = 9.65NNTWDD373 pKa = 3.54GKK375 pKa = 10.9ADD377 pKa = 3.75GSAKK381 pKa = 9.56TLPEE385 pKa = 3.5GTYY388 pKa = 10.11FYY390 pKa = 10.74ILDD393 pKa = 4.72LGNGSPKK400 pKa = 9.9TSGWLYY406 pKa = 11.17LKK408 pKa = 10.73
Molecular weight: 44.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R7L974|A0A2R7L974_9FLAO Peptide chain release factor 3 (Fragment) OS=Flavobacterium sp. HMWF030 OX=2056864 GN=prfC PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1334036 |
16 |
2144 |
281.4 |
31.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.518 ± 0.038 | 0.761 ± 0.012 |
5.37 ± 0.027 | 6.209 ± 0.04 |
5.361 ± 0.029 | 6.232 ± 0.036 |
1.632 ± 0.015 | 8.154 ± 0.036 |
8.197 ± 0.041 | 9.181 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.103 ± 0.019 | 6.425 ± 0.04 |
3.414 ± 0.02 | 3.419 ± 0.021 |
3.133 ± 0.021 | 6.629 ± 0.031 |
5.938 ± 0.043 | 6.13 ± 0.028 |
1.077 ± 0.013 | 4.115 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
