
Olive latent virus 2 (isolate Italy) (OLV-2)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Oleavirus; Olive latent virus 2
Average proteome isoelectric point is 7.83
Get precalculated fractions of proteins
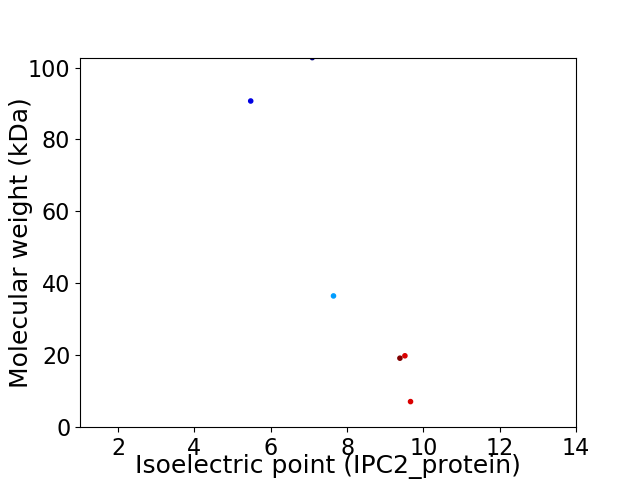
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
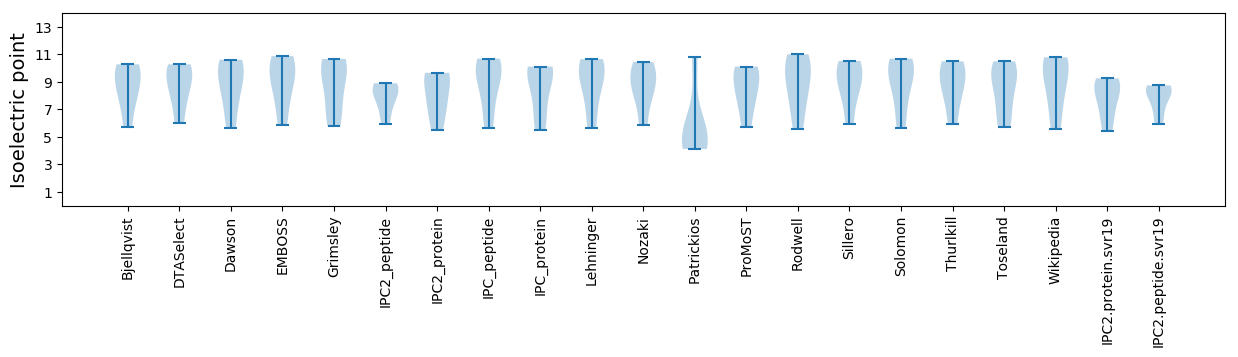
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q83945|Q83945_OLV2I Uncharacterized protein orf1 OS=Olive latent virus 2 (isolate Italy) OX=650489 GN=orf1 PE=4 SV=1
MM1 pKa = 7.61AGVLYY6 pKa = 10.65DD7 pKa = 3.77EE8 pKa = 5.59EE9 pKa = 4.8YY10 pKa = 11.18VPQEE14 pKa = 4.01PDD16 pKa = 3.27YY17 pKa = 11.09LYY19 pKa = 10.92CPYY22 pKa = 9.92EE23 pKa = 4.12WEE25 pKa = 5.04GEE27 pKa = 4.17DD28 pKa = 4.85PTQCDD33 pKa = 3.34ANVPSVSYY41 pKa = 10.55LVEE44 pKa = 3.97AVKK47 pKa = 10.74KK48 pKa = 10.71SFTEE52 pKa = 4.54GNHH55 pKa = 6.77DD56 pKa = 3.69MLISCEE62 pKa = 4.23SYY64 pKa = 10.44HH65 pKa = 7.13AEE67 pKa = 4.16CCEE70 pKa = 3.92GPLMNKK76 pKa = 9.87GLTLSTYY83 pKa = 9.81FMSYY87 pKa = 10.38PGISLPAFGGGVIHH101 pKa = 5.6VTSARR106 pKa = 11.84VCMQAFYY113 pKa = 11.03AVTPGRR119 pKa = 11.84CSCSCGTQFIYY130 pKa = 10.22RR131 pKa = 11.84AKK133 pKa = 10.47GGTGLTFDD141 pKa = 3.98ATNTISSAFKK151 pKa = 10.99FEE153 pKa = 4.47DD154 pKa = 4.42GKK156 pKa = 11.28VKK158 pKa = 10.15WLTEE162 pKa = 4.02DD163 pKa = 3.92LGLALLAGCFLKK175 pKa = 10.77YY176 pKa = 10.77VPIPHH181 pKa = 6.94EE182 pKa = 3.88IEE184 pKa = 3.62IKK186 pKa = 10.1IEE188 pKa = 4.08RR189 pKa = 11.84KK190 pKa = 9.67VEE192 pKa = 3.85PVSALDD198 pKa = 3.39FWEE201 pKa = 4.52PEE203 pKa = 3.91GLISDD208 pKa = 3.61VHH210 pKa = 5.88FVHH213 pKa = 6.32PQEE216 pKa = 4.4RR217 pKa = 11.84ALAPEE222 pKa = 4.29PPMDD226 pKa = 3.53VEE228 pKa = 4.35MVRR231 pKa = 11.84LVRR234 pKa = 11.84TEE236 pKa = 3.9LFEE239 pKa = 4.57HH240 pKa = 6.78FNPTLLLNARR250 pKa = 11.84SDD252 pKa = 3.8FSGEE256 pKa = 3.8KK257 pKa = 10.06LKK259 pKa = 10.86EE260 pKa = 4.02VEE262 pKa = 4.13LPRR265 pKa = 11.84AQSEE269 pKa = 4.23PLVPRR274 pKa = 11.84VCLPRR279 pKa = 11.84GDD281 pKa = 4.97DD282 pKa = 3.52ALIDD286 pKa = 3.36QFFIDD291 pKa = 4.5KK292 pKa = 10.79APEE295 pKa = 3.95HH296 pKa = 6.53FNRR299 pKa = 11.84EE300 pKa = 3.88PATDD304 pKa = 3.27QWQVEE309 pKa = 4.42SDD311 pKa = 3.91DD312 pKa = 3.86FTVDD316 pKa = 3.78FPTQVRR322 pKa = 11.84LRR324 pKa = 11.84EE325 pKa = 3.85RR326 pKa = 11.84DD327 pKa = 3.42EE328 pKa = 4.03DD329 pKa = 3.55WFRR332 pKa = 11.84RR333 pKa = 11.84KK334 pKa = 9.55PDD336 pKa = 3.57FAVSSIRR343 pKa = 11.84TGLEE347 pKa = 3.46MPRR350 pKa = 11.84IATKK354 pKa = 10.78KK355 pKa = 8.46EE356 pKa = 3.36MLMAFSKK363 pKa = 10.83RR364 pKa = 11.84NVGAPQMVEE373 pKa = 4.18VPQEE377 pKa = 3.68IHH379 pKa = 6.05VKK381 pKa = 10.19RR382 pKa = 11.84LADD385 pKa = 3.5RR386 pKa = 11.84FFRR389 pKa = 11.84TFLKK393 pKa = 10.69EE394 pKa = 4.07GGWDD398 pKa = 3.57KK399 pKa = 11.8GDD401 pKa = 3.03GHH403 pKa = 6.72YY404 pKa = 10.32WDD406 pKa = 5.82DD407 pKa = 3.55FFHH410 pKa = 6.95RR411 pKa = 11.84KK412 pKa = 7.49GRR414 pKa = 11.84KK415 pKa = 8.71VFSLEE420 pKa = 3.58NCYY423 pKa = 10.9DD424 pKa = 4.15DD425 pKa = 5.14PLSSYY430 pKa = 11.43LVMLKK435 pKa = 10.42SAGKK439 pKa = 10.22FSLDD443 pKa = 2.97LNFPVRR449 pKa = 11.84SVPQTITYY457 pKa = 8.75HH458 pKa = 5.57EE459 pKa = 4.19KK460 pKa = 9.41WVTEE464 pKa = 4.02VFSPMFLQVMARR476 pKa = 11.84FFSRR480 pKa = 11.84LQDD483 pKa = 3.43WVVVPAGPMADD494 pKa = 4.91FSRR497 pKa = 11.84VWPSFEE503 pKa = 4.27GKK505 pKa = 10.17CLTEE509 pKa = 4.77IDD511 pKa = 3.93LSKK514 pKa = 10.64FDD516 pKa = 3.92KK517 pKa = 11.2SQGKK521 pKa = 8.89LLHH524 pKa = 6.67DD525 pKa = 3.74VQRR528 pKa = 11.84EE529 pKa = 3.63IFLRR533 pKa = 11.84LGFPPIWCDD542 pKa = 2.18WWFKK546 pKa = 9.99FHH548 pKa = 7.01EE549 pKa = 4.35SSYY552 pKa = 11.54LNDD555 pKa = 3.73KK556 pKa = 10.64NLGIAFSVDD565 pKa = 3.14YY566 pKa = 10.6QRR568 pKa = 11.84RR569 pKa = 11.84TGNTATYY576 pKa = 9.93FGNTLVTMIMMAEE589 pKa = 4.2VYY591 pKa = 10.08EE592 pKa = 4.4LDD594 pKa = 3.64SLPFCGLFSGDD605 pKa = 4.24DD606 pKa = 3.63NLIVTDD612 pKa = 4.35RR613 pKa = 11.84PLEE616 pKa = 4.15GRR618 pKa = 11.84VSLFPNMFNMEE629 pKa = 4.64AKK631 pKa = 10.31VMRR634 pKa = 11.84PGQNYY639 pKa = 7.12MCSKK643 pKa = 10.01YY644 pKa = 10.74LCTVDD649 pKa = 4.98GRR651 pKa = 11.84VTPVPDD657 pKa = 3.86PFKK660 pKa = 10.82LLKK663 pKa = 10.22KK664 pKa = 10.48AGASCPLEE672 pKa = 4.36HH673 pKa = 7.35LDD675 pKa = 5.16DD676 pKa = 4.88FYY678 pKa = 11.78EE679 pKa = 4.66SFLDD683 pKa = 3.54YY684 pKa = 10.71TKK686 pKa = 11.2YY687 pKa = 10.53LVRR690 pKa = 11.84DD691 pKa = 3.78EE692 pKa = 4.33VRR694 pKa = 11.84TLIPRR699 pKa = 11.84LMAHH703 pKa = 7.71RR704 pKa = 11.84YY705 pKa = 6.78QVSMDD710 pKa = 3.62EE711 pKa = 4.65AGVALDD717 pKa = 5.63QIMSWRR723 pKa = 11.84KK724 pKa = 9.8SKK726 pKa = 8.67TQFFKK731 pKa = 10.63HH732 pKa = 4.67FQYY735 pKa = 10.72RR736 pKa = 11.84KK737 pKa = 8.84PIKK740 pKa = 10.32DD741 pKa = 3.13SLEE744 pKa = 3.96AVKK747 pKa = 10.75SDD749 pKa = 3.4FLSAFGKK756 pKa = 9.59VDD758 pKa = 3.09DD759 pKa = 5.69RR760 pKa = 11.84IRR762 pKa = 11.84VWRR765 pKa = 11.84GRR767 pKa = 11.84RR768 pKa = 11.84TNHH771 pKa = 6.02KK772 pKa = 10.54KK773 pKa = 10.23EE774 pKa = 3.63VDD776 pKa = 3.42ALKK779 pKa = 10.94VPLVSFRR786 pKa = 11.84EE787 pKa = 4.17
MM1 pKa = 7.61AGVLYY6 pKa = 10.65DD7 pKa = 3.77EE8 pKa = 5.59EE9 pKa = 4.8YY10 pKa = 11.18VPQEE14 pKa = 4.01PDD16 pKa = 3.27YY17 pKa = 11.09LYY19 pKa = 10.92CPYY22 pKa = 9.92EE23 pKa = 4.12WEE25 pKa = 5.04GEE27 pKa = 4.17DD28 pKa = 4.85PTQCDD33 pKa = 3.34ANVPSVSYY41 pKa = 10.55LVEE44 pKa = 3.97AVKK47 pKa = 10.74KK48 pKa = 10.71SFTEE52 pKa = 4.54GNHH55 pKa = 6.77DD56 pKa = 3.69MLISCEE62 pKa = 4.23SYY64 pKa = 10.44HH65 pKa = 7.13AEE67 pKa = 4.16CCEE70 pKa = 3.92GPLMNKK76 pKa = 9.87GLTLSTYY83 pKa = 9.81FMSYY87 pKa = 10.38PGISLPAFGGGVIHH101 pKa = 5.6VTSARR106 pKa = 11.84VCMQAFYY113 pKa = 11.03AVTPGRR119 pKa = 11.84CSCSCGTQFIYY130 pKa = 10.22RR131 pKa = 11.84AKK133 pKa = 10.47GGTGLTFDD141 pKa = 3.98ATNTISSAFKK151 pKa = 10.99FEE153 pKa = 4.47DD154 pKa = 4.42GKK156 pKa = 11.28VKK158 pKa = 10.15WLTEE162 pKa = 4.02DD163 pKa = 3.92LGLALLAGCFLKK175 pKa = 10.77YY176 pKa = 10.77VPIPHH181 pKa = 6.94EE182 pKa = 3.88IEE184 pKa = 3.62IKK186 pKa = 10.1IEE188 pKa = 4.08RR189 pKa = 11.84KK190 pKa = 9.67VEE192 pKa = 3.85PVSALDD198 pKa = 3.39FWEE201 pKa = 4.52PEE203 pKa = 3.91GLISDD208 pKa = 3.61VHH210 pKa = 5.88FVHH213 pKa = 6.32PQEE216 pKa = 4.4RR217 pKa = 11.84ALAPEE222 pKa = 4.29PPMDD226 pKa = 3.53VEE228 pKa = 4.35MVRR231 pKa = 11.84LVRR234 pKa = 11.84TEE236 pKa = 3.9LFEE239 pKa = 4.57HH240 pKa = 6.78FNPTLLLNARR250 pKa = 11.84SDD252 pKa = 3.8FSGEE256 pKa = 3.8KK257 pKa = 10.06LKK259 pKa = 10.86EE260 pKa = 4.02VEE262 pKa = 4.13LPRR265 pKa = 11.84AQSEE269 pKa = 4.23PLVPRR274 pKa = 11.84VCLPRR279 pKa = 11.84GDD281 pKa = 4.97DD282 pKa = 3.52ALIDD286 pKa = 3.36QFFIDD291 pKa = 4.5KK292 pKa = 10.79APEE295 pKa = 3.95HH296 pKa = 6.53FNRR299 pKa = 11.84EE300 pKa = 3.88PATDD304 pKa = 3.27QWQVEE309 pKa = 4.42SDD311 pKa = 3.91DD312 pKa = 3.86FTVDD316 pKa = 3.78FPTQVRR322 pKa = 11.84LRR324 pKa = 11.84EE325 pKa = 3.85RR326 pKa = 11.84DD327 pKa = 3.42EE328 pKa = 4.03DD329 pKa = 3.55WFRR332 pKa = 11.84RR333 pKa = 11.84KK334 pKa = 9.55PDD336 pKa = 3.57FAVSSIRR343 pKa = 11.84TGLEE347 pKa = 3.46MPRR350 pKa = 11.84IATKK354 pKa = 10.78KK355 pKa = 8.46EE356 pKa = 3.36MLMAFSKK363 pKa = 10.83RR364 pKa = 11.84NVGAPQMVEE373 pKa = 4.18VPQEE377 pKa = 3.68IHH379 pKa = 6.05VKK381 pKa = 10.19RR382 pKa = 11.84LADD385 pKa = 3.5RR386 pKa = 11.84FFRR389 pKa = 11.84TFLKK393 pKa = 10.69EE394 pKa = 4.07GGWDD398 pKa = 3.57KK399 pKa = 11.8GDD401 pKa = 3.03GHH403 pKa = 6.72YY404 pKa = 10.32WDD406 pKa = 5.82DD407 pKa = 3.55FFHH410 pKa = 6.95RR411 pKa = 11.84KK412 pKa = 7.49GRR414 pKa = 11.84KK415 pKa = 8.71VFSLEE420 pKa = 3.58NCYY423 pKa = 10.9DD424 pKa = 4.15DD425 pKa = 5.14PLSSYY430 pKa = 11.43LVMLKK435 pKa = 10.42SAGKK439 pKa = 10.22FSLDD443 pKa = 2.97LNFPVRR449 pKa = 11.84SVPQTITYY457 pKa = 8.75HH458 pKa = 5.57EE459 pKa = 4.19KK460 pKa = 9.41WVTEE464 pKa = 4.02VFSPMFLQVMARR476 pKa = 11.84FFSRR480 pKa = 11.84LQDD483 pKa = 3.43WVVVPAGPMADD494 pKa = 4.91FSRR497 pKa = 11.84VWPSFEE503 pKa = 4.27GKK505 pKa = 10.17CLTEE509 pKa = 4.77IDD511 pKa = 3.93LSKK514 pKa = 10.64FDD516 pKa = 3.92KK517 pKa = 11.2SQGKK521 pKa = 8.89LLHH524 pKa = 6.67DD525 pKa = 3.74VQRR528 pKa = 11.84EE529 pKa = 3.63IFLRR533 pKa = 11.84LGFPPIWCDD542 pKa = 2.18WWFKK546 pKa = 9.99FHH548 pKa = 7.01EE549 pKa = 4.35SSYY552 pKa = 11.54LNDD555 pKa = 3.73KK556 pKa = 10.64NLGIAFSVDD565 pKa = 3.14YY566 pKa = 10.6QRR568 pKa = 11.84RR569 pKa = 11.84TGNTATYY576 pKa = 9.93FGNTLVTMIMMAEE589 pKa = 4.2VYY591 pKa = 10.08EE592 pKa = 4.4LDD594 pKa = 3.64SLPFCGLFSGDD605 pKa = 4.24DD606 pKa = 3.63NLIVTDD612 pKa = 4.35RR613 pKa = 11.84PLEE616 pKa = 4.15GRR618 pKa = 11.84VSLFPNMFNMEE629 pKa = 4.64AKK631 pKa = 10.31VMRR634 pKa = 11.84PGQNYY639 pKa = 7.12MCSKK643 pKa = 10.01YY644 pKa = 10.74LCTVDD649 pKa = 4.98GRR651 pKa = 11.84VTPVPDD657 pKa = 3.86PFKK660 pKa = 10.82LLKK663 pKa = 10.22KK664 pKa = 10.48AGASCPLEE672 pKa = 4.36HH673 pKa = 7.35LDD675 pKa = 5.16DD676 pKa = 4.88FYY678 pKa = 11.78EE679 pKa = 4.66SFLDD683 pKa = 3.54YY684 pKa = 10.71TKK686 pKa = 11.2YY687 pKa = 10.53LVRR690 pKa = 11.84DD691 pKa = 3.78EE692 pKa = 4.33VRR694 pKa = 11.84TLIPRR699 pKa = 11.84LMAHH703 pKa = 7.71RR704 pKa = 11.84YY705 pKa = 6.78QVSMDD710 pKa = 3.62EE711 pKa = 4.65AGVALDD717 pKa = 5.63QIMSWRR723 pKa = 11.84KK724 pKa = 9.8SKK726 pKa = 8.67TQFFKK731 pKa = 10.63HH732 pKa = 4.67FQYY735 pKa = 10.72RR736 pKa = 11.84KK737 pKa = 8.84PIKK740 pKa = 10.32DD741 pKa = 3.13SLEE744 pKa = 3.96AVKK747 pKa = 10.75SDD749 pKa = 3.4FLSAFGKK756 pKa = 9.59VDD758 pKa = 3.09DD759 pKa = 5.69RR760 pKa = 11.84IRR762 pKa = 11.84VWRR765 pKa = 11.84GRR767 pKa = 11.84RR768 pKa = 11.84TNHH771 pKa = 6.02KK772 pKa = 10.54KK773 pKa = 10.23EE774 pKa = 3.63VDD776 pKa = 3.42ALKK779 pKa = 10.94VPLVSFRR786 pKa = 11.84EE787 pKa = 4.17
Molecular weight: 90.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q83946|Q83946_OLV2I Capsid protein OS=Olive latent virus 2 (isolate Italy) OX=650489 GN=orf2 PE=4 SV=1
MM1 pKa = 7.57SSGDD5 pKa = 3.45EE6 pKa = 4.17TVPKK10 pKa = 10.38LKK12 pKa = 10.17QSKK15 pKa = 10.26LSLDD19 pKa = 3.17RR20 pKa = 11.84HH21 pKa = 5.45KK22 pKa = 11.16ASWSYY27 pKa = 10.74SVPWQRR33 pKa = 11.84GRR35 pKa = 11.84RR36 pKa = 11.84FRR38 pKa = 11.84VQTSHH43 pKa = 6.9VGAEE47 pKa = 4.04SQYY50 pKa = 9.33PLLVATSSLLLSGAA64 pKa = 3.95
MM1 pKa = 7.57SSGDD5 pKa = 3.45EE6 pKa = 4.17TVPKK10 pKa = 10.38LKK12 pKa = 10.17QSKK15 pKa = 10.26LSLDD19 pKa = 3.17RR20 pKa = 11.84HH21 pKa = 5.45KK22 pKa = 11.16ASWSYY27 pKa = 10.74SVPWQRR33 pKa = 11.84GRR35 pKa = 11.84RR36 pKa = 11.84FRR38 pKa = 11.84VQTSHH43 pKa = 6.9VGAEE47 pKa = 4.04SQYY50 pKa = 9.33PLLVATSSLLLSGAA64 pKa = 3.95
Molecular weight: 7.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2453 |
64 |
908 |
408.8 |
46.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.194 ± 1.143 | 1.508 ± 0.598 |
5.503 ± 0.787 | 6.319 ± 0.622 |
5.544 ± 0.713 | 6.36 ± 1.016 |
2.731 ± 0.281 | 3.384 ± 0.319 |
5.3 ± 1.477 | 8.806 ± 0.535 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.609 ± 0.436 | 2.65 ± 0.183 |
5.096 ± 0.64 | 3.506 ± 0.378 |
6.767 ± 0.975 | 8.194 ± 1.102 |
4.77 ± 0.282 | 7.949 ± 0.876 |
1.631 ± 0.315 | 3.18 ± 0.376 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
