
Rhodospira trueperi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Rhodospira
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
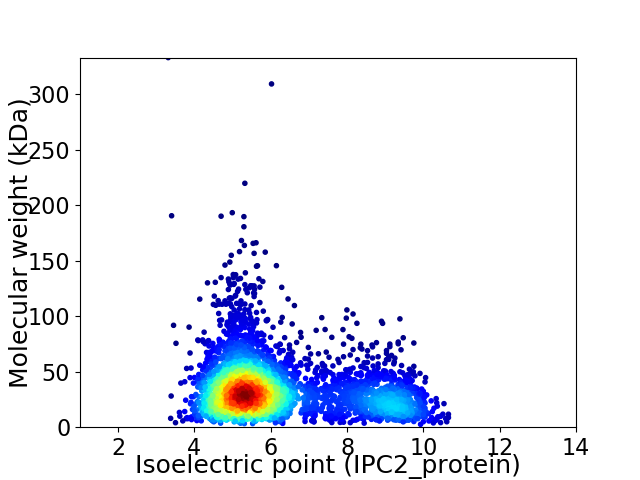
Virtual 2D-PAGE plot for 3690 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G6XJA0|A0A1G6XJA0_9PROT UDP-N-acetylmuramyl pentapeptide phosphotransferase/UDP-N-acetylglucosamine-1-phosphate transferase OS=Rhodospira trueperi OX=69960 GN=SAMN05421720_101526 PE=4 SV=1
MM1 pKa = 7.34SVLAKK6 pKa = 9.84TLTRR10 pKa = 11.84GVAAGALALGLGISTAAIADD30 pKa = 3.73DD31 pKa = 4.52LEE33 pKa = 4.6PGKK36 pKa = 10.68QYY38 pKa = 11.43DD39 pKa = 3.71LGRR42 pKa = 11.84AATAEE47 pKa = 4.55EE48 pKa = 4.04IAGWDD53 pKa = 3.54IDD55 pKa = 4.09VMPDD59 pKa = 3.03GTGAPVGQGTPIEE72 pKa = 4.66GEE74 pKa = 4.31DD75 pKa = 4.33IYY77 pKa = 10.97LTQCASCHH85 pKa = 5.83GEE87 pKa = 3.82FGEE90 pKa = 5.16GMDD93 pKa = 4.5RR94 pKa = 11.84YY95 pKa = 9.36PVLFGGEE102 pKa = 4.09DD103 pKa = 3.35TLASHH108 pKa = 6.8NPVKK112 pKa = 10.22TPGSYY117 pKa = 8.65WPYY120 pKa = 10.85ASTLFDD126 pKa = 4.37YY127 pKa = 10.52IHH129 pKa = 7.09RR130 pKa = 11.84AMPFGQAQTLTNDD143 pKa = 2.94QVYY146 pKa = 10.9ALTAFLLYY154 pKa = 10.4INFVLDD160 pKa = 3.59DD161 pKa = 4.19DD162 pKa = 5.45AFVLSNEE169 pKa = 4.31NIGEE173 pKa = 3.96IEE175 pKa = 4.07MPNRR179 pKa = 11.84DD180 pKa = 3.45GFIPDD185 pKa = 4.9DD186 pKa = 4.45RR187 pKa = 11.84PDD189 pKa = 3.73AQPEE193 pKa = 4.44TVCMSDD199 pKa = 3.45CDD201 pKa = 3.66VGTEE205 pKa = 4.29VIGRR209 pKa = 11.84AAIIDD214 pKa = 3.86VTPDD218 pKa = 2.96GDD220 pKa = 4.01GQPEE224 pKa = 4.88GISLEE229 pKa = 4.15
MM1 pKa = 7.34SVLAKK6 pKa = 9.84TLTRR10 pKa = 11.84GVAAGALALGLGISTAAIADD30 pKa = 3.73DD31 pKa = 4.52LEE33 pKa = 4.6PGKK36 pKa = 10.68QYY38 pKa = 11.43DD39 pKa = 3.71LGRR42 pKa = 11.84AATAEE47 pKa = 4.55EE48 pKa = 4.04IAGWDD53 pKa = 3.54IDD55 pKa = 4.09VMPDD59 pKa = 3.03GTGAPVGQGTPIEE72 pKa = 4.66GEE74 pKa = 4.31DD75 pKa = 4.33IYY77 pKa = 10.97LTQCASCHH85 pKa = 5.83GEE87 pKa = 3.82FGEE90 pKa = 5.16GMDD93 pKa = 4.5RR94 pKa = 11.84YY95 pKa = 9.36PVLFGGEE102 pKa = 4.09DD103 pKa = 3.35TLASHH108 pKa = 6.8NPVKK112 pKa = 10.22TPGSYY117 pKa = 8.65WPYY120 pKa = 10.85ASTLFDD126 pKa = 4.37YY127 pKa = 10.52IHH129 pKa = 7.09RR130 pKa = 11.84AMPFGQAQTLTNDD143 pKa = 2.94QVYY146 pKa = 10.9ALTAFLLYY154 pKa = 10.4INFVLDD160 pKa = 3.59DD161 pKa = 4.19DD162 pKa = 5.45AFVLSNEE169 pKa = 4.31NIGEE173 pKa = 3.96IEE175 pKa = 4.07MPNRR179 pKa = 11.84DD180 pKa = 3.45GFIPDD185 pKa = 4.9DD186 pKa = 4.45RR187 pKa = 11.84PDD189 pKa = 3.73AQPEE193 pKa = 4.44TVCMSDD199 pKa = 3.45CDD201 pKa = 3.66VGTEE205 pKa = 4.29VIGRR209 pKa = 11.84AAIIDD214 pKa = 3.86VTPDD218 pKa = 2.96GDD220 pKa = 4.01GQPEE224 pKa = 4.88GISLEE229 pKa = 4.15
Molecular weight: 24.33 kDa
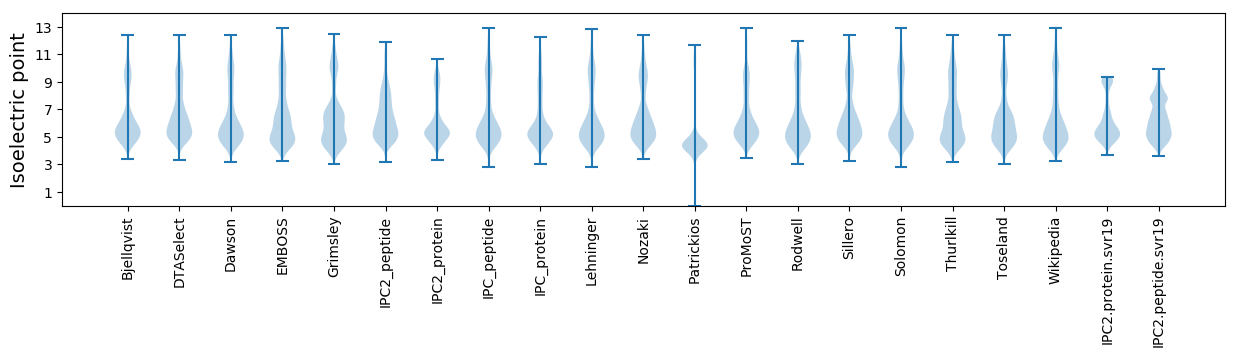
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G7EL61|A0A1G7EL61_9PROT Uncharacterized protein OS=Rhodospira trueperi OX=69960 GN=SAMN05421720_109118 PE=4 SV=1
MM1 pKa = 7.25SRR3 pKa = 11.84RR4 pKa = 11.84AAVYY8 pKa = 10.28LRR10 pKa = 11.84VSTTGQTTEE19 pKa = 3.79NQRR22 pKa = 11.84LEE24 pKa = 4.38LEE26 pKa = 4.34AVAARR31 pKa = 11.84CGWSIIGVYY40 pKa = 9.86EE41 pKa = 3.97DD42 pKa = 4.58AGISGAKK49 pKa = 10.13GRR51 pKa = 11.84DD52 pKa = 3.17KK53 pKa = 10.98RR54 pKa = 11.84PEE56 pKa = 3.67FDD58 pKa = 3.54RR59 pKa = 11.84LCRR62 pKa = 11.84DD63 pKa = 3.08AARR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84FDD70 pKa = 4.43IVMAWSVDD78 pKa = 3.2RR79 pKa = 11.84LGRR82 pKa = 11.84SLQDD86 pKa = 3.11LVAFLGDD93 pKa = 3.21LHH95 pKa = 8.42ASGTHH100 pKa = 6.95LYY102 pKa = 10.3LHH104 pKa = 5.79QQGIDD109 pKa = 3.13TTTPSGKK116 pKa = 10.61AMFQMLGVFAEE127 pKa = 4.92FEE129 pKa = 3.75RR130 pKa = 11.84AMIRR134 pKa = 11.84EE135 pKa = 4.1RR136 pKa = 11.84VQAGLARR143 pKa = 11.84ARR145 pKa = 11.84AQGKK149 pKa = 9.25RR150 pKa = 11.84LGRR153 pKa = 11.84PRR155 pKa = 11.84TPSTTEE161 pKa = 3.63LAIKK165 pKa = 10.4ADD167 pKa = 3.71LRR169 pKa = 11.84AGSMGIRR176 pKa = 11.84KK177 pKa = 8.8IAAKK181 pKa = 9.93HH182 pKa = 4.66GVGVSVVQRR191 pKa = 11.84LRR193 pKa = 11.84AGMDD197 pKa = 3.18SDD199 pKa = 4.47VVV201 pKa = 3.44
MM1 pKa = 7.25SRR3 pKa = 11.84RR4 pKa = 11.84AAVYY8 pKa = 10.28LRR10 pKa = 11.84VSTTGQTTEE19 pKa = 3.79NQRR22 pKa = 11.84LEE24 pKa = 4.38LEE26 pKa = 4.34AVAARR31 pKa = 11.84CGWSIIGVYY40 pKa = 9.86EE41 pKa = 3.97DD42 pKa = 4.58AGISGAKK49 pKa = 10.13GRR51 pKa = 11.84DD52 pKa = 3.17KK53 pKa = 10.98RR54 pKa = 11.84PEE56 pKa = 3.67FDD58 pKa = 3.54RR59 pKa = 11.84LCRR62 pKa = 11.84DD63 pKa = 3.08AARR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84FDD70 pKa = 4.43IVMAWSVDD78 pKa = 3.2RR79 pKa = 11.84LGRR82 pKa = 11.84SLQDD86 pKa = 3.11LVAFLGDD93 pKa = 3.21LHH95 pKa = 8.42ASGTHH100 pKa = 6.95LYY102 pKa = 10.3LHH104 pKa = 5.79QQGIDD109 pKa = 3.13TTTPSGKK116 pKa = 10.61AMFQMLGVFAEE127 pKa = 4.92FEE129 pKa = 3.75RR130 pKa = 11.84AMIRR134 pKa = 11.84EE135 pKa = 4.1RR136 pKa = 11.84VQAGLARR143 pKa = 11.84ARR145 pKa = 11.84AQGKK149 pKa = 9.25RR150 pKa = 11.84LGRR153 pKa = 11.84PRR155 pKa = 11.84TPSTTEE161 pKa = 3.63LAIKK165 pKa = 10.4ADD167 pKa = 3.71LRR169 pKa = 11.84AGSMGIRR176 pKa = 11.84KK177 pKa = 8.8IAAKK181 pKa = 9.93HH182 pKa = 4.66GVGVSVVQRR191 pKa = 11.84LRR193 pKa = 11.84AGMDD197 pKa = 3.18SDD199 pKa = 4.47VVV201 pKa = 3.44
Molecular weight: 22.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1220506 |
24 |
3194 |
330.8 |
35.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.807 ± 0.064 | 0.877 ± 0.012 |
6.485 ± 0.034 | 5.729 ± 0.038 |
3.248 ± 0.027 | 8.841 ± 0.042 |
2.214 ± 0.02 | 4.268 ± 0.031 |
2.326 ± 0.031 | 10.406 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.502 ± 0.018 | 2.176 ± 0.021 |
5.783 ± 0.039 | 2.696 ± 0.023 |
7.954 ± 0.046 | 4.771 ± 0.029 |
5.665 ± 0.033 | 7.935 ± 0.034 |
1.386 ± 0.017 | 1.931 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
