
Maribacter vaceletii
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Maribacter
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
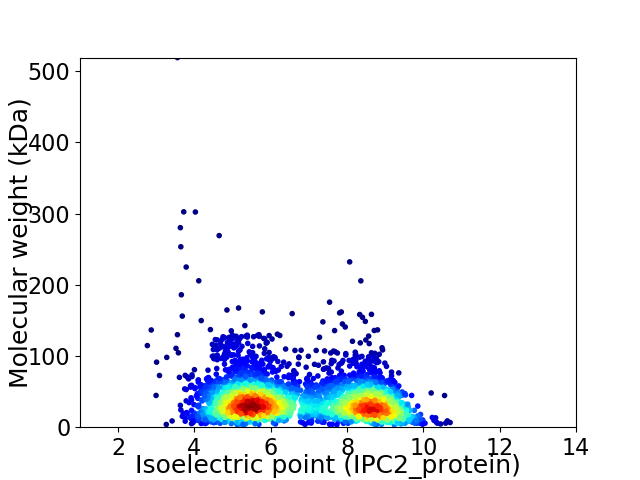
Virtual 2D-PAGE plot for 3317 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A495E6A9|A0A495E6A9_9FLAO Uncharacterized protein OS=Maribacter vaceletii OX=1206816 GN=CLV91_2581 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.44KK3 pKa = 8.44ITNTKK8 pKa = 7.68YY9 pKa = 10.41VKK11 pKa = 10.68NIFASALMLVATTSTFAQDD30 pKa = 2.98EE31 pKa = 4.39EE32 pKa = 4.46AAEE35 pKa = 4.08EE36 pKa = 4.15APKK39 pKa = 11.0LEE41 pKa = 4.03ISGSVDD47 pKa = 2.63AYY49 pKa = 11.2YY50 pKa = 9.01RR51 pKa = 11.84TAFNDD56 pKa = 3.24AGVASTDD63 pKa = 3.38PGTAFANQTGFALGMGNLIASYY85 pKa = 10.91DD86 pKa = 3.64AGKK89 pKa = 10.66VGAVLDD95 pKa = 3.89IVVGPRR101 pKa = 11.84GAGATFNTDD110 pKa = 2.52IVDD113 pKa = 4.88GIVNQAYY120 pKa = 9.75VYY122 pKa = 10.42WNVSDD127 pKa = 3.83KK128 pKa = 8.92TTLTMGRR135 pKa = 11.84FNTFLGYY142 pKa = 10.2EE143 pKa = 4.37VIAPQANFNYY153 pKa = 8.18STSYY157 pKa = 10.84LFSNGPFSHH166 pKa = 6.72MGLKK170 pKa = 10.22ADD172 pKa = 4.86FALSDD177 pKa = 4.42DD178 pKa = 3.89FSLMVGVMNPWDD190 pKa = 3.78TNDD193 pKa = 2.92ISGTGEE199 pKa = 3.95YY200 pKa = 10.75SFGAQLGFYY209 pKa = 9.73GQYY212 pKa = 11.08LNFYY216 pKa = 9.24YY217 pKa = 10.59DD218 pKa = 3.09SGKK221 pKa = 10.5NGGLGTEE228 pKa = 3.96IDD230 pKa = 3.8FTGGFDD236 pKa = 3.32ISDD239 pKa = 3.55AFFLGINAAYY249 pKa = 7.5DD250 pKa = 3.68TNDD253 pKa = 3.38AADD256 pKa = 3.32TGFYY260 pKa = 10.66GVALYY265 pKa = 8.64PQIATSDD272 pKa = 3.37AFSIGLRR279 pKa = 11.84GEE281 pKa = 4.39YY282 pKa = 9.46FASTIDD288 pKa = 4.14GGDD291 pKa = 3.41NYY293 pKa = 11.51DD294 pKa = 4.3DD295 pKa = 3.58VTAITLTGSYY305 pKa = 10.59SVDD308 pKa = 3.1NLILKK313 pKa = 10.13PEE315 pKa = 3.99FRR317 pKa = 11.84LDD319 pKa = 3.75SIKK322 pKa = 11.17DD323 pKa = 3.48GFTDD327 pKa = 3.34SDD329 pKa = 3.94GAASDD334 pKa = 3.84SLATFALAAIYY345 pKa = 10.58SFF347 pKa = 4.19
MM1 pKa = 7.61KK2 pKa = 10.44KK3 pKa = 8.44ITNTKK8 pKa = 7.68YY9 pKa = 10.41VKK11 pKa = 10.68NIFASALMLVATTSTFAQDD30 pKa = 2.98EE31 pKa = 4.39EE32 pKa = 4.46AAEE35 pKa = 4.08EE36 pKa = 4.15APKK39 pKa = 11.0LEE41 pKa = 4.03ISGSVDD47 pKa = 2.63AYY49 pKa = 11.2YY50 pKa = 9.01RR51 pKa = 11.84TAFNDD56 pKa = 3.24AGVASTDD63 pKa = 3.38PGTAFANQTGFALGMGNLIASYY85 pKa = 10.91DD86 pKa = 3.64AGKK89 pKa = 10.66VGAVLDD95 pKa = 3.89IVVGPRR101 pKa = 11.84GAGATFNTDD110 pKa = 2.52IVDD113 pKa = 4.88GIVNQAYY120 pKa = 9.75VYY122 pKa = 10.42WNVSDD127 pKa = 3.83KK128 pKa = 8.92TTLTMGRR135 pKa = 11.84FNTFLGYY142 pKa = 10.2EE143 pKa = 4.37VIAPQANFNYY153 pKa = 8.18STSYY157 pKa = 10.84LFSNGPFSHH166 pKa = 6.72MGLKK170 pKa = 10.22ADD172 pKa = 4.86FALSDD177 pKa = 4.42DD178 pKa = 3.89FSLMVGVMNPWDD190 pKa = 3.78TNDD193 pKa = 2.92ISGTGEE199 pKa = 3.95YY200 pKa = 10.75SFGAQLGFYY209 pKa = 9.73GQYY212 pKa = 11.08LNFYY216 pKa = 9.24YY217 pKa = 10.59DD218 pKa = 3.09SGKK221 pKa = 10.5NGGLGTEE228 pKa = 3.96IDD230 pKa = 3.8FTGGFDD236 pKa = 3.32ISDD239 pKa = 3.55AFFLGINAAYY249 pKa = 7.5DD250 pKa = 3.68TNDD253 pKa = 3.38AADD256 pKa = 3.32TGFYY260 pKa = 10.66GVALYY265 pKa = 8.64PQIATSDD272 pKa = 3.37AFSIGLRR279 pKa = 11.84GEE281 pKa = 4.39YY282 pKa = 9.46FASTIDD288 pKa = 4.14GGDD291 pKa = 3.41NYY293 pKa = 11.51DD294 pKa = 4.3DD295 pKa = 3.58VTAITLTGSYY305 pKa = 10.59SVDD308 pKa = 3.1NLILKK313 pKa = 10.13PEE315 pKa = 3.99FRR317 pKa = 11.84LDD319 pKa = 3.75SIKK322 pKa = 11.17DD323 pKa = 3.48GFTDD327 pKa = 3.34SDD329 pKa = 3.94GAASDD334 pKa = 3.84SLATFALAAIYY345 pKa = 10.58SFF347 pKa = 4.19
Molecular weight: 36.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A495EC27|A0A495EC27_9FLAO Thioredoxin OS=Maribacter vaceletii OX=1206816 GN=CLV91_0510 PE=3 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.32AITADD27 pKa = 3.83KK28 pKa = 10.62PEE30 pKa = 4.36KK31 pKa = 10.61SLLAPLKK38 pKa = 10.66KK39 pKa = 10.41SGGRR43 pKa = 11.84NSQGKK48 pKa = 5.87MTMRR52 pKa = 11.84HH53 pKa = 5.73RR54 pKa = 11.84GGGHH58 pKa = 5.08KK59 pKa = 9.74RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.28RR63 pKa = 11.84VIDD66 pKa = 4.1FKK68 pKa = 11.12RR69 pKa = 11.84DD70 pKa = 3.57KK71 pKa = 10.71QDD73 pKa = 2.61VAATVKK79 pKa = 10.68SIEE82 pKa = 4.02YY83 pKa = 10.15DD84 pKa = 3.43PNRR87 pKa = 11.84TAFIALLEE95 pKa = 4.26YY96 pKa = 10.99ADD98 pKa = 4.12GVKK101 pKa = 10.19KK102 pKa = 10.6YY103 pKa = 10.48IIAQNGLQVGQKK115 pKa = 9.74VSSGQGAAPEE125 pKa = 4.24VGNALALSEE134 pKa = 4.55IPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGTFAQLMAKK167 pKa = 9.93DD168 pKa = 4.08GKK170 pKa = 10.42FVTVKK175 pKa = 9.72MPSGEE180 pKa = 4.06TRR182 pKa = 11.84LILSGCMATIGAISNSDD199 pKa = 3.23HH200 pKa = 6.21QLLVSGKK207 pKa = 9.87AGRR210 pKa = 11.84TRR212 pKa = 11.84WLGKK216 pKa = 9.9RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.36GFRR256 pKa = 11.84TRR258 pKa = 11.84SEE260 pKa = 4.05TKK262 pKa = 9.05STNKK266 pKa = 10.46YY267 pKa = 9.26IIEE270 pKa = 4.03RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.32AITADD27 pKa = 3.83KK28 pKa = 10.62PEE30 pKa = 4.36KK31 pKa = 10.61SLLAPLKK38 pKa = 10.66KK39 pKa = 10.41SGGRR43 pKa = 11.84NSQGKK48 pKa = 5.87MTMRR52 pKa = 11.84HH53 pKa = 5.73RR54 pKa = 11.84GGGHH58 pKa = 5.08KK59 pKa = 9.74RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.28RR63 pKa = 11.84VIDD66 pKa = 4.1FKK68 pKa = 11.12RR69 pKa = 11.84DD70 pKa = 3.57KK71 pKa = 10.71QDD73 pKa = 2.61VAATVKK79 pKa = 10.68SIEE82 pKa = 4.02YY83 pKa = 10.15DD84 pKa = 3.43PNRR87 pKa = 11.84TAFIALLEE95 pKa = 4.26YY96 pKa = 10.99ADD98 pKa = 4.12GVKK101 pKa = 10.19KK102 pKa = 10.6YY103 pKa = 10.48IIAQNGLQVGQKK115 pKa = 9.74VSSGQGAAPEE125 pKa = 4.24VGNALALSEE134 pKa = 4.55IPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAVMARR156 pKa = 11.84SAGTFAQLMAKK167 pKa = 9.93DD168 pKa = 4.08GKK170 pKa = 10.42FVTVKK175 pKa = 9.72MPSGEE180 pKa = 4.06TRR182 pKa = 11.84LILSGCMATIGAISNSDD199 pKa = 3.23HH200 pKa = 6.21QLLVSGKK207 pKa = 9.87AGRR210 pKa = 11.84TRR212 pKa = 11.84WLGKK216 pKa = 9.9RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.36GFRR256 pKa = 11.84TRR258 pKa = 11.84SEE260 pKa = 4.05TKK262 pKa = 9.05STNKK266 pKa = 10.46YY267 pKa = 9.26IIEE270 pKa = 4.03RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
Molecular weight: 29.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1184146 |
30 |
4889 |
357.0 |
40.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.251 ± 0.042 | 0.741 ± 0.015 |
5.394 ± 0.041 | 6.682 ± 0.046 |
5.082 ± 0.032 | 6.567 ± 0.044 |
1.718 ± 0.018 | 8.027 ± 0.042 |
8.329 ± 0.067 | 9.111 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.096 ± 0.023 | 6.464 ± 0.045 |
3.418 ± 0.024 | 3.028 ± 0.021 |
3.144 ± 0.029 | 6.527 ± 0.031 |
6.01 ± 0.057 | 6.196 ± 0.031 |
1.134 ± 0.017 | 4.083 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
