
Rabbit hemorrhagic disease virus (strain Rabbit/Germany/FRG/1989) (Ra/LV/RHDV/GH/1989/GE) (RHDV-FRG)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Picornavirales; Caliciviridae; Lagovirus; Rabbit hemorrhagic disease virus
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
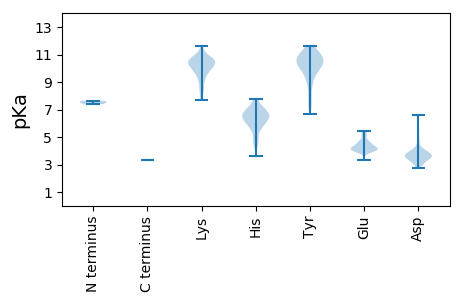
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P27410-2|POLG-2_RHDVF Isoform of P27410 Isoform Subgenomic capsid protein VP60 of Genome polyprotein OS=Rabbit hemorrhagic disease virus (strain Rabbit/Germany/FRG/1989) OX=314536 GN=ORF1 PE=1 SV=1
MM1 pKa = 7.39EE2 pKa = 5.14GKK4 pKa = 10.19ARR6 pKa = 11.84AAPQGEE12 pKa = 4.33AAGTATTASVPGTTTDD28 pKa = 3.0GMDD31 pKa = 3.74PGVVATTSVITAEE44 pKa = 4.19NSSASIATAGIGGPPQQVDD63 pKa = 3.54QQEE66 pKa = 4.05TWRR69 pKa = 11.84TNFYY73 pKa = 10.86YY74 pKa = 10.89NDD76 pKa = 3.15VFTWSVADD84 pKa = 3.75APGSILYY91 pKa = 7.84TVQHH95 pKa = 6.35SPQNNPFTAVLSQMYY110 pKa = 9.86AGWAGGMQFRR120 pKa = 11.84FIVAGSGVFGGRR132 pKa = 11.84LVRR135 pKa = 11.84AVIPPGIEE143 pKa = 3.36IGPGLEE149 pKa = 3.93VRR151 pKa = 11.84QFPHH155 pKa = 6.28VVIDD159 pKa = 3.75ARR161 pKa = 11.84SLEE164 pKa = 4.3PVTITMPDD172 pKa = 3.5LRR174 pKa = 11.84PNMYY178 pKa = 10.15HH179 pKa = 6.3PTGDD183 pKa = 3.64PGLVPTLVLSVYY195 pKa = 10.89NNLINPFGGSTSAIQVTVEE214 pKa = 4.06TRR216 pKa = 11.84PSEE219 pKa = 4.15DD220 pKa = 3.43FEE222 pKa = 4.67FVMIRR227 pKa = 11.84APSSKK232 pKa = 9.04TVDD235 pKa = 3.84SISPAGLLTTPVLTGVGNDD254 pKa = 3.6NRR256 pKa = 11.84WNGQIVGLQPVPGGFSTCNRR276 pKa = 11.84HH277 pKa = 4.62WNLNGSTYY285 pKa = 10.49GWSSPRR291 pKa = 11.84FGDD294 pKa = 2.95IDD296 pKa = 3.54HH297 pKa = 7.02RR298 pKa = 11.84RR299 pKa = 11.84GSASYY304 pKa = 10.46SGSNATNVLQFWYY317 pKa = 10.18ANAGSAIDD325 pKa = 3.98NPISQVAPDD334 pKa = 4.89GFPDD338 pKa = 3.45MSFVPFNGPGIPAAGWVGFGAIWNSNSGAPNVTTVQAYY376 pKa = 9.18EE377 pKa = 4.06LGFATGAPGNLQPTTNTSGAQTVAKK402 pKa = 9.9SIYY405 pKa = 10.6AVVTGTAQNPAGLFVMASGIISTPNASAITYY436 pKa = 7.48TPQPDD441 pKa = 3.88RR442 pKa = 11.84IVTTPGTPAAAPVGKK457 pKa = 8.48NTPIMFASVVRR468 pKa = 11.84RR469 pKa = 11.84TGDD472 pKa = 3.2VNATAGSANGTQYY485 pKa = 10.31GTGSQPLPVTIGLSLNNYY503 pKa = 9.5SSALMPGQFFVWQLTFASGFMEE525 pKa = 4.67IGLSVDD531 pKa = 3.57GYY533 pKa = 10.65FYY535 pKa = 11.2AGTGASTTLIDD546 pKa = 3.95LTEE549 pKa = 5.34LIDD552 pKa = 3.77VRR554 pKa = 11.84PVGPRR559 pKa = 11.84PSKK562 pKa = 9.82STLVFNLGGTANGFSYY578 pKa = 11.12VV579 pKa = 3.31
MM1 pKa = 7.39EE2 pKa = 5.14GKK4 pKa = 10.19ARR6 pKa = 11.84AAPQGEE12 pKa = 4.33AAGTATTASVPGTTTDD28 pKa = 3.0GMDD31 pKa = 3.74PGVVATTSVITAEE44 pKa = 4.19NSSASIATAGIGGPPQQVDD63 pKa = 3.54QQEE66 pKa = 4.05TWRR69 pKa = 11.84TNFYY73 pKa = 10.86YY74 pKa = 10.89NDD76 pKa = 3.15VFTWSVADD84 pKa = 3.75APGSILYY91 pKa = 7.84TVQHH95 pKa = 6.35SPQNNPFTAVLSQMYY110 pKa = 9.86AGWAGGMQFRR120 pKa = 11.84FIVAGSGVFGGRR132 pKa = 11.84LVRR135 pKa = 11.84AVIPPGIEE143 pKa = 3.36IGPGLEE149 pKa = 3.93VRR151 pKa = 11.84QFPHH155 pKa = 6.28VVIDD159 pKa = 3.75ARR161 pKa = 11.84SLEE164 pKa = 4.3PVTITMPDD172 pKa = 3.5LRR174 pKa = 11.84PNMYY178 pKa = 10.15HH179 pKa = 6.3PTGDD183 pKa = 3.64PGLVPTLVLSVYY195 pKa = 10.89NNLINPFGGSTSAIQVTVEE214 pKa = 4.06TRR216 pKa = 11.84PSEE219 pKa = 4.15DD220 pKa = 3.43FEE222 pKa = 4.67FVMIRR227 pKa = 11.84APSSKK232 pKa = 9.04TVDD235 pKa = 3.84SISPAGLLTTPVLTGVGNDD254 pKa = 3.6NRR256 pKa = 11.84WNGQIVGLQPVPGGFSTCNRR276 pKa = 11.84HH277 pKa = 4.62WNLNGSTYY285 pKa = 10.49GWSSPRR291 pKa = 11.84FGDD294 pKa = 2.95IDD296 pKa = 3.54HH297 pKa = 7.02RR298 pKa = 11.84RR299 pKa = 11.84GSASYY304 pKa = 10.46SGSNATNVLQFWYY317 pKa = 10.18ANAGSAIDD325 pKa = 3.98NPISQVAPDD334 pKa = 4.89GFPDD338 pKa = 3.45MSFVPFNGPGIPAAGWVGFGAIWNSNSGAPNVTTVQAYY376 pKa = 9.18EE377 pKa = 4.06LGFATGAPGNLQPTTNTSGAQTVAKK402 pKa = 9.9SIYY405 pKa = 10.6AVVTGTAQNPAGLFVMASGIISTPNASAITYY436 pKa = 7.48TPQPDD441 pKa = 3.88RR442 pKa = 11.84IVTTPGTPAAAPVGKK457 pKa = 8.48NTPIMFASVVRR468 pKa = 11.84RR469 pKa = 11.84TGDD472 pKa = 3.2VNATAGSANGTQYY485 pKa = 10.31GTGSQPLPVTIGLSLNNYY503 pKa = 9.5SSALMPGQFFVWQLTFASGFMEE525 pKa = 4.67IGLSVDD531 pKa = 3.57GYY533 pKa = 10.65FYY535 pKa = 11.2AGTGASTTLIDD546 pKa = 3.95LTEE549 pKa = 5.34LIDD552 pKa = 3.77VRR554 pKa = 11.84PVGPRR559 pKa = 11.84PSKK562 pKa = 9.82STLVFNLGGTANGFSYY578 pKa = 11.12VV579 pKa = 3.31
Molecular weight: 60.26 kDa
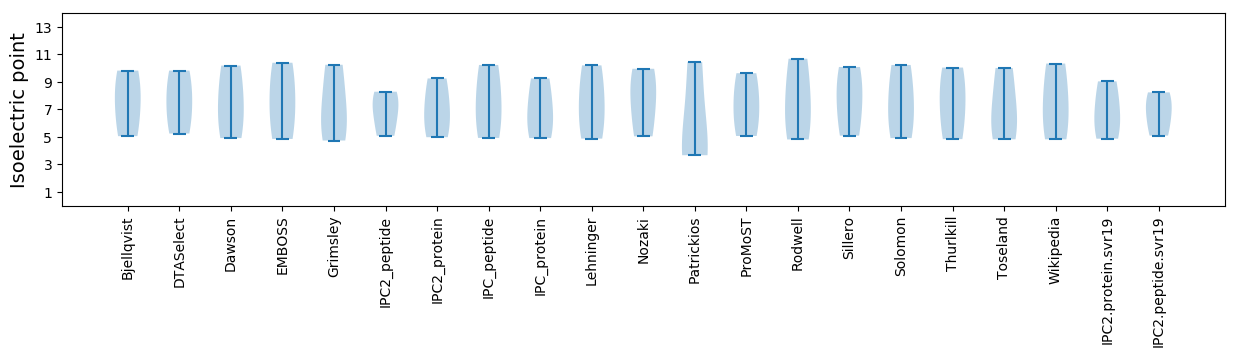
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P27410-2|POLG-2_RHDVF Isoform of P27410 Isoform Subgenomic capsid protein VP60 of Genome polyprotein OS=Rabbit hemorrhagic disease virus (strain Rabbit/Germany/FRG/1989) OX=314536 GN=ORF1 PE=1 SV=1
MM1 pKa = 7.64AFLMSEE7 pKa = 4.62FIGLGLAGAGVLSNALLRR25 pKa = 11.84RR26 pKa = 11.84QEE28 pKa = 4.08LQLQKK33 pKa = 10.83QAMEE37 pKa = 4.22NGLVLKK43 pKa = 10.49ADD45 pKa = 3.38QLGRR49 pKa = 11.84LGFNPNEE56 pKa = 4.18VKK58 pKa = 10.77NVIVGNSFSSNVRR71 pKa = 11.84LSNMHH76 pKa = 6.52NDD78 pKa = 3.1ASVVNAYY85 pKa = 9.78NVYY88 pKa = 10.11NPASNGIRR96 pKa = 11.84KK97 pKa = 9.52KK98 pKa = 10.48IKK100 pKa = 10.21SLNNSVKK107 pKa = 10.06IYY109 pKa = 8.66NTTGEE114 pKa = 4.24SSVV117 pKa = 3.3
MM1 pKa = 7.64AFLMSEE7 pKa = 4.62FIGLGLAGAGVLSNALLRR25 pKa = 11.84RR26 pKa = 11.84QEE28 pKa = 4.08LQLQKK33 pKa = 10.83QAMEE37 pKa = 4.22NGLVLKK43 pKa = 10.49ADD45 pKa = 3.38QLGRR49 pKa = 11.84LGFNPNEE56 pKa = 4.18VKK58 pKa = 10.77NVIVGNSFSSNVRR71 pKa = 11.84LSNMHH76 pKa = 6.52NDD78 pKa = 3.1ASVVNAYY85 pKa = 9.78NVYY88 pKa = 10.11NPASNGIRR96 pKa = 11.84KK97 pKa = 9.52KK98 pKa = 10.48IKK100 pKa = 10.21SLNNSVKK107 pKa = 10.06IYY109 pKa = 8.66NTTGEE114 pKa = 4.24SSVV117 pKa = 3.3
Molecular weight: 12.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3040 |
117 |
2344 |
1013.3 |
110.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.961 ± 0.662 | 1.842 ± 0.893 |
5.164 ± 1.155 | 3.52 ± 0.484 |
4.375 ± 0.222 | 8.75 ± 1.241 |
1.678 ± 0.423 | 5.066 ± 0.18 |
4.375 ± 1.331 | 7.763 ± 1.203 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.467 ± 0.234 | 5.362 ± 2.041 |
5.822 ± 1.205 | 3.651 ± 0.277 |
4.276 ± 0.326 | 6.974 ± 1.106 |
7.895 ± 1.447 | 8.52 ± 0.227 |
1.546 ± 0.353 | 2.993 ± 0.111 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
