
Kosmotoga sp.
Taxonomy: cellular organisms; Bacteria; Thermotogae; Thermotogae; Kosmotogales; Kosmotogaceae; Kosmotoga; unclassified Kosmotoga
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
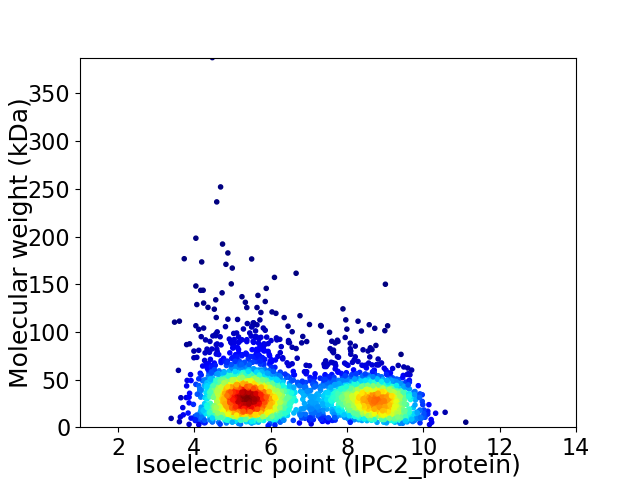
Virtual 2D-PAGE plot for 2971 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5D0TAZ2|A0A5D0TAZ2_9BACT B12-binding domain-containing radical SAM protein OS=Kosmotoga sp. OX=1955248 GN=FXF54_00930 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 10.35KK3 pKa = 10.46LLILSTIIVAVMALISCTATMNILPKK29 pKa = 10.5SILIDD34 pKa = 4.04DD35 pKa = 3.9SHH37 pKa = 7.71NNQFGYY43 pKa = 10.55VDD45 pKa = 3.89EE46 pKa = 5.28NFFATMINTLEE57 pKa = 4.27GEE59 pKa = 4.29NYY61 pKa = 9.93QVDD64 pKa = 4.21FTSDD68 pKa = 3.02VGFSPDD74 pKa = 3.35QYY76 pKa = 11.3DD77 pKa = 3.38ILIIPTPVSLYY88 pKa = 11.02SNTEE92 pKa = 3.46INQVKK97 pKa = 10.62SFVQNGNKK105 pKa = 9.77IIVLGEE111 pKa = 3.82FYY113 pKa = 10.66SYY115 pKa = 11.51YY116 pKa = 10.96DD117 pKa = 3.57NSEE120 pKa = 3.94LNYY123 pKa = 10.22ILNQVGADD131 pKa = 3.49MQFDD135 pKa = 3.57NSLITDD141 pKa = 3.89SVNNYY146 pKa = 9.91DD147 pKa = 6.13NNDD150 pKa = 3.17EE151 pKa = 4.1WPEE154 pKa = 3.64ISLFTPHH161 pKa = 7.14PTTDD165 pKa = 3.31SLNSIVLFAASSLNVFGDD183 pKa = 4.15AIKK186 pKa = 10.32IAEE189 pKa = 4.22SSSINLVEE197 pKa = 4.48TFDD200 pKa = 3.94SSTFNEE206 pKa = 4.41SGLKK210 pKa = 9.97AGLMEE215 pKa = 4.35NQDD218 pKa = 3.78YY219 pKa = 11.15DD220 pKa = 4.05SQTTAYY226 pKa = 10.13VSVAAVDD233 pKa = 3.95KK234 pKa = 10.78IGNGKK239 pKa = 8.83VVAVTDD245 pKa = 3.73VNLFSDD251 pKa = 4.4DD252 pKa = 3.41VDD254 pKa = 4.66DD255 pKa = 5.95FFPSLDD261 pKa = 3.47SMLNVADD268 pKa = 4.62NKK270 pKa = 11.03DD271 pKa = 3.53FLINIVNWW279 pKa = 3.5
MM1 pKa = 7.63KK2 pKa = 10.35KK3 pKa = 10.46LLILSTIIVAVMALISCTATMNILPKK29 pKa = 10.5SILIDD34 pKa = 4.04DD35 pKa = 3.9SHH37 pKa = 7.71NNQFGYY43 pKa = 10.55VDD45 pKa = 3.89EE46 pKa = 5.28NFFATMINTLEE57 pKa = 4.27GEE59 pKa = 4.29NYY61 pKa = 9.93QVDD64 pKa = 4.21FTSDD68 pKa = 3.02VGFSPDD74 pKa = 3.35QYY76 pKa = 11.3DD77 pKa = 3.38ILIIPTPVSLYY88 pKa = 11.02SNTEE92 pKa = 3.46INQVKK97 pKa = 10.62SFVQNGNKK105 pKa = 9.77IIVLGEE111 pKa = 3.82FYY113 pKa = 10.66SYY115 pKa = 11.51YY116 pKa = 10.96DD117 pKa = 3.57NSEE120 pKa = 3.94LNYY123 pKa = 10.22ILNQVGADD131 pKa = 3.49MQFDD135 pKa = 3.57NSLITDD141 pKa = 3.89SVNNYY146 pKa = 9.91DD147 pKa = 6.13NNDD150 pKa = 3.17EE151 pKa = 4.1WPEE154 pKa = 3.64ISLFTPHH161 pKa = 7.14PTTDD165 pKa = 3.31SLNSIVLFAASSLNVFGDD183 pKa = 4.15AIKK186 pKa = 10.32IAEE189 pKa = 4.22SSSINLVEE197 pKa = 4.48TFDD200 pKa = 3.94SSTFNEE206 pKa = 4.41SGLKK210 pKa = 9.97AGLMEE215 pKa = 4.35NQDD218 pKa = 3.78YY219 pKa = 11.15DD220 pKa = 4.05SQTTAYY226 pKa = 10.13VSVAAVDD233 pKa = 3.95KK234 pKa = 10.78IGNGKK239 pKa = 8.83VVAVTDD245 pKa = 3.73VNLFSDD251 pKa = 4.4DD252 pKa = 3.41VDD254 pKa = 4.66DD255 pKa = 5.95FFPSLDD261 pKa = 3.47SMLNVADD268 pKa = 4.62NKK270 pKa = 11.03DD271 pKa = 3.53FLINIVNWW279 pKa = 3.5
Molecular weight: 30.91 kDa
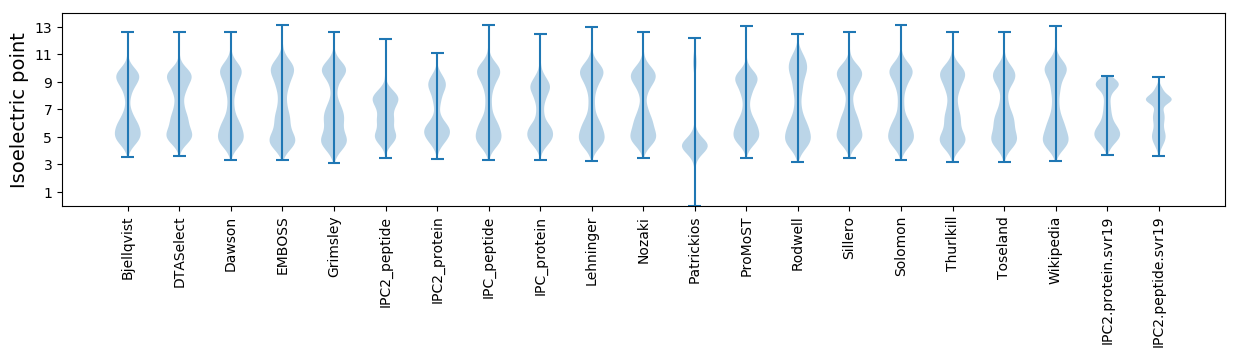
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5D0SFV1|A0A5D0SFV1_9BACT 30S ribosomal protein S18 OS=Kosmotoga sp. OX=1955248 GN=rpsR PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.18QPSRR9 pKa = 11.84SKK11 pKa = 10.76RR12 pKa = 11.84KK13 pKa = 8.13RR14 pKa = 11.84THH16 pKa = 6.16GFLARR21 pKa = 11.84KK22 pKa = 6.9KK23 pKa = 7.78TKK25 pKa = 9.6AGKK28 pKa = 10.12RR29 pKa = 11.84ILANRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 10.0KK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 9.07RR41 pKa = 11.84IAAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.18QPSRR9 pKa = 11.84SKK11 pKa = 10.76RR12 pKa = 11.84KK13 pKa = 8.13RR14 pKa = 11.84THH16 pKa = 6.16GFLARR21 pKa = 11.84KK22 pKa = 6.9KK23 pKa = 7.78TKK25 pKa = 9.6AGKK28 pKa = 10.12RR29 pKa = 11.84ILANRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 10.0KK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 9.07RR41 pKa = 11.84IAAA44 pKa = 3.93
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
932970 |
13 |
3461 |
314.0 |
35.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.535 ± 0.04 | 0.792 ± 0.016 |
5.443 ± 0.038 | 7.796 ± 0.049 |
5.097 ± 0.04 | 6.471 ± 0.033 |
1.57 ± 0.017 | 8.699 ± 0.034 |
8.177 ± 0.064 | 9.521 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.432 ± 0.02 | 5.358 ± 0.033 |
3.521 ± 0.026 | 2.227 ± 0.018 |
4.103 ± 0.032 | 6.525 ± 0.036 |
5.087 ± 0.038 | 6.566 ± 0.031 |
1.021 ± 0.018 | 4.059 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
