
Drosophila A virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
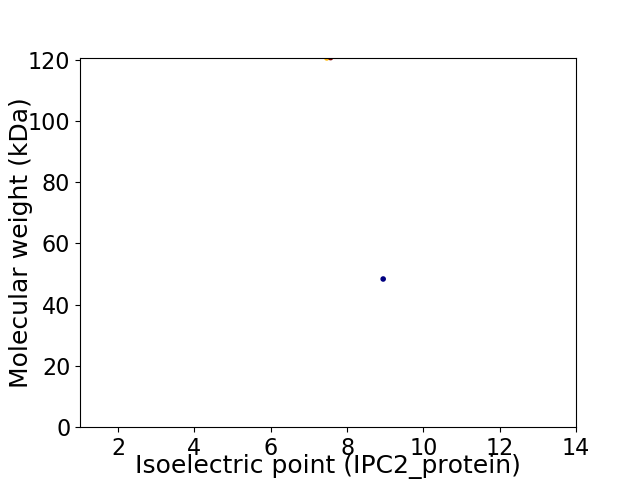
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6FGJ6|C6FGJ6_9VIRU Capsid protein OS=Drosophila A virus OX=595895 PE=3 SV=1
MM1 pKa = 7.89DD2 pKa = 5.63ASNPIISSDD11 pKa = 3.13RR12 pKa = 11.84STVNQVYY19 pKa = 10.8AKK21 pKa = 8.34MVRR24 pKa = 11.84RR25 pKa = 11.84KK26 pKa = 9.91RR27 pKa = 11.84QDD29 pKa = 3.14LAYY32 pKa = 9.99LQEE35 pKa = 4.49LVRR38 pKa = 11.84TAHH41 pKa = 7.52RR42 pKa = 11.84EE43 pKa = 3.66ILPQGEE49 pKa = 4.6LPHH52 pKa = 6.55EE53 pKa = 4.74SEE55 pKa = 4.14AKK57 pKa = 10.34QYY59 pKa = 9.04VAEE62 pKa = 4.26YY63 pKa = 9.9KK64 pKa = 10.69ARR66 pKa = 11.84LPVVDD71 pKa = 3.8TKK73 pKa = 11.25KK74 pKa = 10.75VGEE77 pKa = 4.42LPTEE81 pKa = 4.42VFQVALPSIPVRR93 pKa = 11.84GIMDD97 pKa = 4.02AVGVPLHH104 pKa = 6.6PPSINNRR111 pKa = 11.84GGHH114 pKa = 4.3THH116 pKa = 5.49VTRR119 pKa = 11.84QMGGVPHH126 pKa = 7.35PPLGLIPIAQKK137 pKa = 10.91YY138 pKa = 8.65GVEE141 pKa = 4.08ILDD144 pKa = 3.53QKK146 pKa = 10.86HH147 pKa = 6.03VYY149 pKa = 8.85TGGTWDD155 pKa = 3.84GFLSRR160 pKa = 11.84LAQQMGRR167 pKa = 11.84TTVPLARR174 pKa = 11.84TVSAGKK180 pKa = 10.43KK181 pKa = 8.46NASDD185 pKa = 3.42RR186 pKa = 11.84AIIKK190 pKa = 9.68LLNKK194 pKa = 9.81YY195 pKa = 7.53MPRR198 pKa = 11.84TGNKK202 pKa = 8.54SWPGVHH208 pKa = 5.45TTPEE212 pKa = 3.9EE213 pKa = 4.79GILDD217 pKa = 4.91GIKK220 pKa = 8.85ITAKK224 pKa = 10.21SSAGAPYY231 pKa = 8.86WRR233 pKa = 11.84HH234 pKa = 5.72KK235 pKa = 10.91GEE237 pKa = 4.49CLDD240 pKa = 4.83HH241 pKa = 8.26IIDD244 pKa = 3.7TGLPVVLKK252 pKa = 10.22HH253 pKa = 6.23IKK255 pKa = 10.24EE256 pKa = 4.25GTLNQLWRR264 pKa = 11.84EE265 pKa = 4.0NPEE268 pKa = 3.83MFLVEE273 pKa = 4.51VKK275 pKa = 10.91NKK277 pKa = 9.66LDD279 pKa = 3.43RR280 pKa = 11.84YY281 pKa = 9.18EE282 pKa = 3.95VSKK285 pKa = 11.07LKK287 pKa = 10.71EE288 pKa = 3.48KK289 pKa = 9.57TRR291 pKa = 11.84PYY293 pKa = 10.4VCVPAHH299 pKa = 5.87WALLFSCLTQGFQEE313 pKa = 4.45GLQVFSNLDD322 pKa = 3.42PSTEE326 pKa = 3.95CSNAYY331 pKa = 9.84GFSSIAGGLTRR342 pKa = 11.84MVDD345 pKa = 2.61WMYY348 pKa = 11.39ACPKK352 pKa = 9.33RR353 pKa = 11.84RR354 pKa = 11.84GRR356 pKa = 11.84VVCYY360 pKa = 10.63GDD362 pKa = 3.82DD363 pKa = 3.4ACITFWSQGVLYY375 pKa = 10.45RR376 pKa = 11.84VDD378 pKa = 3.88PDD380 pKa = 4.19FKK382 pKa = 11.51QMDD385 pKa = 3.82GSIDD389 pKa = 3.76RR390 pKa = 11.84EE391 pKa = 4.05DD392 pKa = 3.42ARR394 pKa = 11.84ITIEE398 pKa = 3.99WVLHH402 pKa = 5.56HH403 pKa = 6.5LRR405 pKa = 11.84KK406 pKa = 10.16DD407 pKa = 3.37LGVEE411 pKa = 3.84EE412 pKa = 5.43TPAFWKK418 pKa = 9.32TVAAVWLDD426 pKa = 3.59MAIDD430 pKa = 3.33PHH432 pKa = 7.58FIVDD436 pKa = 4.2GKK438 pKa = 8.74TVYY441 pKa = 10.03KK442 pKa = 10.36KK443 pKa = 10.64KK444 pKa = 10.35NPHH447 pKa = 5.6GLMTGVPGTTLFDD460 pKa = 3.7TVKK463 pKa = 10.86SVIVWNEE470 pKa = 3.52MLDD473 pKa = 3.65QASVGSIDD481 pKa = 4.79LLNEE485 pKa = 3.99AQVVEE490 pKa = 4.08WMKK493 pKa = 10.86RR494 pKa = 11.84QGLVVKK500 pKa = 9.81EE501 pKa = 4.52GTWSPVALPARR512 pKa = 11.84DD513 pKa = 3.81TEE515 pKa = 4.8GLITDD520 pKa = 4.16HH521 pKa = 6.99KK522 pKa = 10.45FLGVQIMGVYY532 pKa = 9.12HH533 pKa = 6.02RR534 pKa = 11.84HH535 pKa = 5.26RR536 pKa = 11.84VIHH539 pKa = 6.02VPTMPEE545 pKa = 3.56SDD547 pKa = 4.04ALEE550 pKa = 4.0MMLCQKK556 pKa = 10.76DD557 pKa = 3.78NPFEE561 pKa = 4.23KK562 pKa = 10.44AVSRR566 pKa = 11.84TARR569 pKa = 11.84QRR571 pKa = 11.84TLYY574 pKa = 11.04DD575 pKa = 3.18RR576 pKa = 11.84MRR578 pKa = 11.84GLMITMGFSIPRR590 pKa = 11.84IEE592 pKa = 4.24EE593 pKa = 3.99TIHH596 pKa = 6.74AVVNTIPGEE605 pKa = 4.12IIVMQTQEE613 pKa = 3.84QTGTKK618 pKa = 9.06PEE620 pKa = 5.12HH621 pKa = 6.14ITLQDD626 pKa = 3.06FEE628 pKa = 5.25YY629 pKa = 9.51PDD631 pKa = 3.53SSGFPSRR638 pKa = 11.84DD639 pKa = 3.09FCLDD643 pKa = 3.98LYY645 pKa = 11.57SDD647 pKa = 4.68GEE649 pKa = 4.14DD650 pKa = 5.53DD651 pKa = 3.7KK652 pKa = 11.97AGWINLFPTLSGFLDD667 pKa = 3.51EE668 pKa = 5.73FKK670 pKa = 10.73RR671 pKa = 11.84EE672 pKa = 3.52QRR674 pKa = 11.84VAVRR678 pKa = 11.84QINLTVQSNDD688 pKa = 3.06YY689 pKa = 10.36DD690 pKa = 3.79VKK692 pKa = 10.93EE693 pKa = 4.37VVGCPPPPEE702 pKa = 5.2ANLNDD707 pKa = 3.72EE708 pKa = 4.59YY709 pKa = 11.66KK710 pKa = 10.86VFEE713 pKa = 4.29ALKK716 pKa = 8.54PQQVQYY722 pKa = 10.91SEE724 pKa = 4.42PNPRR728 pKa = 11.84PKK730 pKa = 10.13VVRR733 pKa = 11.84ITEE736 pKa = 4.18SGDD739 pKa = 3.15IPEE742 pKa = 4.64KK743 pKa = 10.4FLPNMAQAVVRR754 pKa = 11.84WLTSVGGVSQVGTVADD770 pKa = 3.72KK771 pKa = 11.3VGASAYY777 pKa = 10.05QIVVGAAKK785 pKa = 10.24GGYY788 pKa = 7.52FTTGDD793 pKa = 3.77EE794 pKa = 4.32LGDD797 pKa = 4.53LISLYY802 pKa = 10.33PLVTPFPTLQDD813 pKa = 3.6SQRR816 pKa = 11.84EE817 pKa = 4.08EE818 pKa = 4.07MEE820 pKa = 4.21EE821 pKa = 3.77KK822 pKa = 10.61RR823 pKa = 11.84NLIDD827 pKa = 3.03KK828 pKa = 7.4TTAARR833 pKa = 11.84TSALRR838 pKa = 11.84RR839 pKa = 11.84EE840 pKa = 4.36IVKK843 pKa = 7.83TQPEE847 pKa = 5.14LINLDD852 pKa = 3.48VAGVSNLHH860 pKa = 6.56PPPHH864 pKa = 7.45DD865 pKa = 3.44INTAEE870 pKa = 4.33DD871 pKa = 3.06AMAYY875 pKa = 7.49VHH877 pKa = 6.8AMASGRR883 pKa = 11.84FSGFTKK889 pKa = 9.95WISEE893 pKa = 4.04VRR895 pKa = 11.84PNAANPVGVRR905 pKa = 11.84LYY907 pKa = 10.75AHH909 pKa = 6.01SHH911 pKa = 5.06HH912 pKa = 6.31TQDD915 pKa = 3.78VKK917 pKa = 11.4LLGEE921 pKa = 4.4AWSSSAKK928 pKa = 9.71LAKK931 pKa = 10.11EE932 pKa = 4.08YY933 pKa = 10.29ICAAILEE940 pKa = 4.73VNNIPYY946 pKa = 9.84KK947 pKa = 10.26KK948 pKa = 10.43SSFATPNVVPPPPEE962 pKa = 4.24SSWARR967 pKa = 11.84QVEE970 pKa = 4.8YY971 pKa = 9.16ATAPKK976 pKa = 9.98AVPQIVPVHH985 pKa = 5.34EE986 pKa = 4.29TLDD989 pKa = 3.7LEE991 pKa = 4.55LFHH994 pKa = 8.92SIMQQFPGHH1003 pKa = 6.69CPRR1006 pKa = 11.84QVQMLIRR1013 pKa = 11.84EE1014 pKa = 4.32VSHH1017 pKa = 6.91KK1018 pKa = 11.14NPDD1021 pKa = 3.54DD1022 pKa = 3.54YY1023 pKa = 10.97RR1024 pKa = 11.84VRR1026 pKa = 11.84IQDD1029 pKa = 3.64LFSRR1033 pKa = 11.84FPPPKK1038 pKa = 9.02VASKK1042 pKa = 10.75RR1043 pKa = 11.84SLLTAEE1049 pKa = 3.93QRR1051 pKa = 11.84TRR1053 pKa = 11.84LNKK1056 pKa = 9.03KK1057 pKa = 7.47TLDD1060 pKa = 3.06RR1061 pKa = 11.84KK1062 pKa = 9.65KK1063 pKa = 10.33RR1064 pKa = 11.84KK1065 pKa = 9.75KK1066 pKa = 9.79IASKK1070 pKa = 10.73LLPTLL1075 pKa = 3.87
MM1 pKa = 7.89DD2 pKa = 5.63ASNPIISSDD11 pKa = 3.13RR12 pKa = 11.84STVNQVYY19 pKa = 10.8AKK21 pKa = 8.34MVRR24 pKa = 11.84RR25 pKa = 11.84KK26 pKa = 9.91RR27 pKa = 11.84QDD29 pKa = 3.14LAYY32 pKa = 9.99LQEE35 pKa = 4.49LVRR38 pKa = 11.84TAHH41 pKa = 7.52RR42 pKa = 11.84EE43 pKa = 3.66ILPQGEE49 pKa = 4.6LPHH52 pKa = 6.55EE53 pKa = 4.74SEE55 pKa = 4.14AKK57 pKa = 10.34QYY59 pKa = 9.04VAEE62 pKa = 4.26YY63 pKa = 9.9KK64 pKa = 10.69ARR66 pKa = 11.84LPVVDD71 pKa = 3.8TKK73 pKa = 11.25KK74 pKa = 10.75VGEE77 pKa = 4.42LPTEE81 pKa = 4.42VFQVALPSIPVRR93 pKa = 11.84GIMDD97 pKa = 4.02AVGVPLHH104 pKa = 6.6PPSINNRR111 pKa = 11.84GGHH114 pKa = 4.3THH116 pKa = 5.49VTRR119 pKa = 11.84QMGGVPHH126 pKa = 7.35PPLGLIPIAQKK137 pKa = 10.91YY138 pKa = 8.65GVEE141 pKa = 4.08ILDD144 pKa = 3.53QKK146 pKa = 10.86HH147 pKa = 6.03VYY149 pKa = 8.85TGGTWDD155 pKa = 3.84GFLSRR160 pKa = 11.84LAQQMGRR167 pKa = 11.84TTVPLARR174 pKa = 11.84TVSAGKK180 pKa = 10.43KK181 pKa = 8.46NASDD185 pKa = 3.42RR186 pKa = 11.84AIIKK190 pKa = 9.68LLNKK194 pKa = 9.81YY195 pKa = 7.53MPRR198 pKa = 11.84TGNKK202 pKa = 8.54SWPGVHH208 pKa = 5.45TTPEE212 pKa = 3.9EE213 pKa = 4.79GILDD217 pKa = 4.91GIKK220 pKa = 8.85ITAKK224 pKa = 10.21SSAGAPYY231 pKa = 8.86WRR233 pKa = 11.84HH234 pKa = 5.72KK235 pKa = 10.91GEE237 pKa = 4.49CLDD240 pKa = 4.83HH241 pKa = 8.26IIDD244 pKa = 3.7TGLPVVLKK252 pKa = 10.22HH253 pKa = 6.23IKK255 pKa = 10.24EE256 pKa = 4.25GTLNQLWRR264 pKa = 11.84EE265 pKa = 4.0NPEE268 pKa = 3.83MFLVEE273 pKa = 4.51VKK275 pKa = 10.91NKK277 pKa = 9.66LDD279 pKa = 3.43RR280 pKa = 11.84YY281 pKa = 9.18EE282 pKa = 3.95VSKK285 pKa = 11.07LKK287 pKa = 10.71EE288 pKa = 3.48KK289 pKa = 9.57TRR291 pKa = 11.84PYY293 pKa = 10.4VCVPAHH299 pKa = 5.87WALLFSCLTQGFQEE313 pKa = 4.45GLQVFSNLDD322 pKa = 3.42PSTEE326 pKa = 3.95CSNAYY331 pKa = 9.84GFSSIAGGLTRR342 pKa = 11.84MVDD345 pKa = 2.61WMYY348 pKa = 11.39ACPKK352 pKa = 9.33RR353 pKa = 11.84RR354 pKa = 11.84GRR356 pKa = 11.84VVCYY360 pKa = 10.63GDD362 pKa = 3.82DD363 pKa = 3.4ACITFWSQGVLYY375 pKa = 10.45RR376 pKa = 11.84VDD378 pKa = 3.88PDD380 pKa = 4.19FKK382 pKa = 11.51QMDD385 pKa = 3.82GSIDD389 pKa = 3.76RR390 pKa = 11.84EE391 pKa = 4.05DD392 pKa = 3.42ARR394 pKa = 11.84ITIEE398 pKa = 3.99WVLHH402 pKa = 5.56HH403 pKa = 6.5LRR405 pKa = 11.84KK406 pKa = 10.16DD407 pKa = 3.37LGVEE411 pKa = 3.84EE412 pKa = 5.43TPAFWKK418 pKa = 9.32TVAAVWLDD426 pKa = 3.59MAIDD430 pKa = 3.33PHH432 pKa = 7.58FIVDD436 pKa = 4.2GKK438 pKa = 8.74TVYY441 pKa = 10.03KK442 pKa = 10.36KK443 pKa = 10.64KK444 pKa = 10.35NPHH447 pKa = 5.6GLMTGVPGTTLFDD460 pKa = 3.7TVKK463 pKa = 10.86SVIVWNEE470 pKa = 3.52MLDD473 pKa = 3.65QASVGSIDD481 pKa = 4.79LLNEE485 pKa = 3.99AQVVEE490 pKa = 4.08WMKK493 pKa = 10.86RR494 pKa = 11.84QGLVVKK500 pKa = 9.81EE501 pKa = 4.52GTWSPVALPARR512 pKa = 11.84DD513 pKa = 3.81TEE515 pKa = 4.8GLITDD520 pKa = 4.16HH521 pKa = 6.99KK522 pKa = 10.45FLGVQIMGVYY532 pKa = 9.12HH533 pKa = 6.02RR534 pKa = 11.84HH535 pKa = 5.26RR536 pKa = 11.84VIHH539 pKa = 6.02VPTMPEE545 pKa = 3.56SDD547 pKa = 4.04ALEE550 pKa = 4.0MMLCQKK556 pKa = 10.76DD557 pKa = 3.78NPFEE561 pKa = 4.23KK562 pKa = 10.44AVSRR566 pKa = 11.84TARR569 pKa = 11.84QRR571 pKa = 11.84TLYY574 pKa = 11.04DD575 pKa = 3.18RR576 pKa = 11.84MRR578 pKa = 11.84GLMITMGFSIPRR590 pKa = 11.84IEE592 pKa = 4.24EE593 pKa = 3.99TIHH596 pKa = 6.74AVVNTIPGEE605 pKa = 4.12IIVMQTQEE613 pKa = 3.84QTGTKK618 pKa = 9.06PEE620 pKa = 5.12HH621 pKa = 6.14ITLQDD626 pKa = 3.06FEE628 pKa = 5.25YY629 pKa = 9.51PDD631 pKa = 3.53SSGFPSRR638 pKa = 11.84DD639 pKa = 3.09FCLDD643 pKa = 3.98LYY645 pKa = 11.57SDD647 pKa = 4.68GEE649 pKa = 4.14DD650 pKa = 5.53DD651 pKa = 3.7KK652 pKa = 11.97AGWINLFPTLSGFLDD667 pKa = 3.51EE668 pKa = 5.73FKK670 pKa = 10.73RR671 pKa = 11.84EE672 pKa = 3.52QRR674 pKa = 11.84VAVRR678 pKa = 11.84QINLTVQSNDD688 pKa = 3.06YY689 pKa = 10.36DD690 pKa = 3.79VKK692 pKa = 10.93EE693 pKa = 4.37VVGCPPPPEE702 pKa = 5.2ANLNDD707 pKa = 3.72EE708 pKa = 4.59YY709 pKa = 11.66KK710 pKa = 10.86VFEE713 pKa = 4.29ALKK716 pKa = 8.54PQQVQYY722 pKa = 10.91SEE724 pKa = 4.42PNPRR728 pKa = 11.84PKK730 pKa = 10.13VVRR733 pKa = 11.84ITEE736 pKa = 4.18SGDD739 pKa = 3.15IPEE742 pKa = 4.64KK743 pKa = 10.4FLPNMAQAVVRR754 pKa = 11.84WLTSVGGVSQVGTVADD770 pKa = 3.72KK771 pKa = 11.3VGASAYY777 pKa = 10.05QIVVGAAKK785 pKa = 10.24GGYY788 pKa = 7.52FTTGDD793 pKa = 3.77EE794 pKa = 4.32LGDD797 pKa = 4.53LISLYY802 pKa = 10.33PLVTPFPTLQDD813 pKa = 3.6SQRR816 pKa = 11.84EE817 pKa = 4.08EE818 pKa = 4.07MEE820 pKa = 4.21EE821 pKa = 3.77KK822 pKa = 10.61RR823 pKa = 11.84NLIDD827 pKa = 3.03KK828 pKa = 7.4TTAARR833 pKa = 11.84TSALRR838 pKa = 11.84RR839 pKa = 11.84EE840 pKa = 4.36IVKK843 pKa = 7.83TQPEE847 pKa = 5.14LINLDD852 pKa = 3.48VAGVSNLHH860 pKa = 6.56PPPHH864 pKa = 7.45DD865 pKa = 3.44INTAEE870 pKa = 4.33DD871 pKa = 3.06AMAYY875 pKa = 7.49VHH877 pKa = 6.8AMASGRR883 pKa = 11.84FSGFTKK889 pKa = 9.95WISEE893 pKa = 4.04VRR895 pKa = 11.84PNAANPVGVRR905 pKa = 11.84LYY907 pKa = 10.75AHH909 pKa = 6.01SHH911 pKa = 5.06HH912 pKa = 6.31TQDD915 pKa = 3.78VKK917 pKa = 11.4LLGEE921 pKa = 4.4AWSSSAKK928 pKa = 9.71LAKK931 pKa = 10.11EE932 pKa = 4.08YY933 pKa = 10.29ICAAILEE940 pKa = 4.73VNNIPYY946 pKa = 9.84KK947 pKa = 10.26KK948 pKa = 10.43SSFATPNVVPPPPEE962 pKa = 4.24SSWARR967 pKa = 11.84QVEE970 pKa = 4.8YY971 pKa = 9.16ATAPKK976 pKa = 9.98AVPQIVPVHH985 pKa = 5.34EE986 pKa = 4.29TLDD989 pKa = 3.7LEE991 pKa = 4.55LFHH994 pKa = 8.92SIMQQFPGHH1003 pKa = 6.69CPRR1006 pKa = 11.84QVQMLIRR1013 pKa = 11.84EE1014 pKa = 4.32VSHH1017 pKa = 6.91KK1018 pKa = 11.14NPDD1021 pKa = 3.54DD1022 pKa = 3.54YY1023 pKa = 10.97RR1024 pKa = 11.84VRR1026 pKa = 11.84IQDD1029 pKa = 3.64LFSRR1033 pKa = 11.84FPPPKK1038 pKa = 9.02VASKK1042 pKa = 10.75RR1043 pKa = 11.84SLLTAEE1049 pKa = 3.93QRR1051 pKa = 11.84TRR1053 pKa = 11.84LNKK1056 pKa = 9.03KK1057 pKa = 7.47TLDD1060 pKa = 3.06RR1061 pKa = 11.84KK1062 pKa = 9.65KK1063 pKa = 10.33RR1064 pKa = 11.84KK1065 pKa = 9.75KK1066 pKa = 9.79IASKK1070 pKa = 10.73LLPTLL1075 pKa = 3.87
Molecular weight: 120.57 kDa
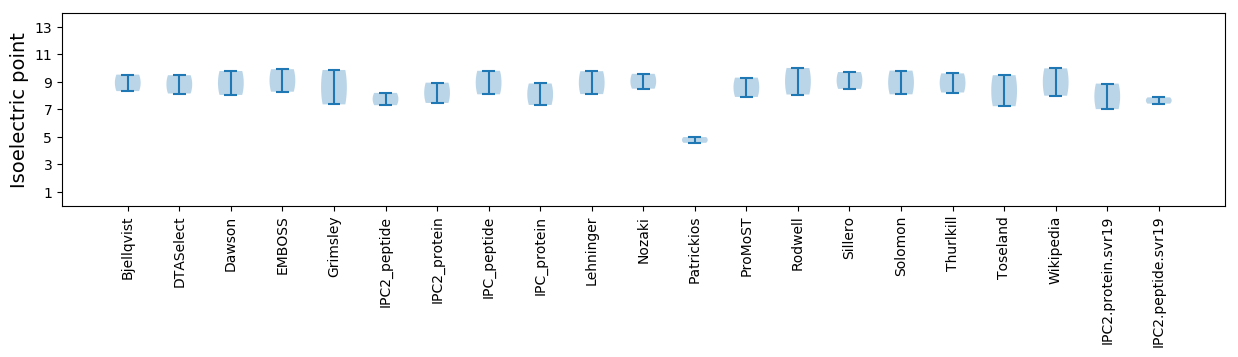
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6FGJ6|C6FGJ6_9VIRU Capsid protein OS=Drosophila A virus OX=595895 PE=3 SV=1
MM1 pKa = 7.09ATIVTTTKK9 pKa = 10.49RR10 pKa = 11.84ISNNSGNNFSTRR22 pKa = 11.84NRR24 pKa = 11.84RR25 pKa = 11.84TPKK28 pKa = 9.22VTVQSVSSRR37 pKa = 11.84QIVGKK42 pKa = 9.52NCPMVQGPRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84GRR55 pKa = 11.84NRR57 pKa = 11.84PRR59 pKa = 11.84KK60 pKa = 9.57SGSQVAYY67 pKa = 10.43SAMPKK72 pKa = 7.91TTLSGVDD79 pKa = 3.34IIGVIEE85 pKa = 4.17VQPQRR90 pKa = 11.84AVGASLFAQAICPTAMSDD108 pKa = 4.99SRR110 pKa = 11.84LQQQSKK116 pKa = 10.42LYY118 pKa = 10.65SRR120 pKa = 11.84WRR122 pKa = 11.84PKK124 pKa = 10.1KK125 pKa = 10.44LRR127 pKa = 11.84VLVVGSGSANTFGSLAIGWSPDD149 pKa = 3.21PANNISGNDD158 pKa = 3.59TQNLNRR164 pKa = 11.84VMACRR169 pKa = 11.84PSSMNRR175 pKa = 11.84MNQTTVLNIPVATTRR190 pKa = 11.84KK191 pKa = 8.93WYY193 pKa = 8.21LTKK196 pKa = 10.91GNPDD200 pKa = 3.67DD201 pKa = 5.25ANHH204 pKa = 6.19GFINVVVASTTGGYY218 pKa = 8.17TGSTTFTVTLEE229 pKa = 3.87WVIEE233 pKa = 4.12FEE235 pKa = 4.14GAEE238 pKa = 4.12LGAVSIDD245 pKa = 3.56EE246 pKa = 6.03DD247 pKa = 3.68ITPDD251 pKa = 3.56TGFQDD256 pKa = 5.11LFTTSDD262 pKa = 3.64GSWNSEE268 pKa = 3.85YY269 pKa = 10.44LTMKK273 pKa = 8.65MHH275 pKa = 7.22HH276 pKa = 6.95GGQMVPFSAAHH287 pKa = 4.42QAYY290 pKa = 9.92VYY292 pKa = 10.43TNAPGTNVTYY302 pKa = 10.7YY303 pKa = 11.08LEE305 pKa = 5.49DD306 pKa = 3.31KK307 pKa = 8.03TTLKK311 pKa = 9.55TANFFAVIQGYY322 pKa = 8.8SIPGFVMFASYY333 pKa = 10.71TDD335 pKa = 3.13AKK337 pKa = 11.04DD338 pKa = 3.58YY339 pKa = 11.16IRR341 pKa = 11.84TGDD344 pKa = 3.12TGKK347 pKa = 10.37CLKK350 pKa = 10.4YY351 pKa = 8.72YY352 pKa = 8.93TQGPVISPPIPKK364 pKa = 9.66FRR366 pKa = 11.84VADD369 pKa = 3.71EE370 pKa = 4.24AVQTVEE376 pKa = 4.01TNLLRR381 pKa = 11.84RR382 pKa = 11.84IDD384 pKa = 3.63QLTEE388 pKa = 3.26QLRR391 pKa = 11.84AVNAPVTRR399 pKa = 11.84TVALSKK405 pKa = 10.09DD406 pKa = 3.27TAKK409 pKa = 10.34IRR411 pKa = 11.84TKK413 pKa = 10.96LEE415 pKa = 3.76GMPDD419 pKa = 3.54GTTLSPFKK427 pKa = 10.79VISEE431 pKa = 4.55DD432 pKa = 3.28PVAAQDD438 pKa = 3.73FEE440 pKa = 4.65IVV442 pKa = 3.15
MM1 pKa = 7.09ATIVTTTKK9 pKa = 10.49RR10 pKa = 11.84ISNNSGNNFSTRR22 pKa = 11.84NRR24 pKa = 11.84RR25 pKa = 11.84TPKK28 pKa = 9.22VTVQSVSSRR37 pKa = 11.84QIVGKK42 pKa = 9.52NCPMVQGPRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84GRR55 pKa = 11.84NRR57 pKa = 11.84PRR59 pKa = 11.84KK60 pKa = 9.57SGSQVAYY67 pKa = 10.43SAMPKK72 pKa = 7.91TTLSGVDD79 pKa = 3.34IIGVIEE85 pKa = 4.17VQPQRR90 pKa = 11.84AVGASLFAQAICPTAMSDD108 pKa = 4.99SRR110 pKa = 11.84LQQQSKK116 pKa = 10.42LYY118 pKa = 10.65SRR120 pKa = 11.84WRR122 pKa = 11.84PKK124 pKa = 10.1KK125 pKa = 10.44LRR127 pKa = 11.84VLVVGSGSANTFGSLAIGWSPDD149 pKa = 3.21PANNISGNDD158 pKa = 3.59TQNLNRR164 pKa = 11.84VMACRR169 pKa = 11.84PSSMNRR175 pKa = 11.84MNQTTVLNIPVATTRR190 pKa = 11.84KK191 pKa = 8.93WYY193 pKa = 8.21LTKK196 pKa = 10.91GNPDD200 pKa = 3.67DD201 pKa = 5.25ANHH204 pKa = 6.19GFINVVVASTTGGYY218 pKa = 8.17TGSTTFTVTLEE229 pKa = 3.87WVIEE233 pKa = 4.12FEE235 pKa = 4.14GAEE238 pKa = 4.12LGAVSIDD245 pKa = 3.56EE246 pKa = 6.03DD247 pKa = 3.68ITPDD251 pKa = 3.56TGFQDD256 pKa = 5.11LFTTSDD262 pKa = 3.64GSWNSEE268 pKa = 3.85YY269 pKa = 10.44LTMKK273 pKa = 8.65MHH275 pKa = 7.22HH276 pKa = 6.95GGQMVPFSAAHH287 pKa = 4.42QAYY290 pKa = 9.92VYY292 pKa = 10.43TNAPGTNVTYY302 pKa = 10.7YY303 pKa = 11.08LEE305 pKa = 5.49DD306 pKa = 3.31KK307 pKa = 8.03TTLKK311 pKa = 9.55TANFFAVIQGYY322 pKa = 8.8SIPGFVMFASYY333 pKa = 10.71TDD335 pKa = 3.13AKK337 pKa = 11.04DD338 pKa = 3.58YY339 pKa = 11.16IRR341 pKa = 11.84TGDD344 pKa = 3.12TGKK347 pKa = 10.37CLKK350 pKa = 10.4YY351 pKa = 8.72YY352 pKa = 8.93TQGPVISPPIPKK364 pKa = 9.66FRR366 pKa = 11.84VADD369 pKa = 3.71EE370 pKa = 4.24AVQTVEE376 pKa = 4.01TNLLRR381 pKa = 11.84RR382 pKa = 11.84IDD384 pKa = 3.63QLTEE388 pKa = 3.26QLRR391 pKa = 11.84AVNAPVTRR399 pKa = 11.84TVALSKK405 pKa = 10.09DD406 pKa = 3.27TAKK409 pKa = 10.34IRR411 pKa = 11.84TKK413 pKa = 10.96LEE415 pKa = 3.76GMPDD419 pKa = 3.54GTTLSPFKK427 pKa = 10.79VISEE431 pKa = 4.55DD432 pKa = 3.28PVAAQDD438 pKa = 3.73FEE440 pKa = 4.65IVV442 pKa = 3.15
Molecular weight: 48.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1517 |
442 |
1075 |
758.5 |
84.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.987 ± 0.243 | 1.055 ± 0.076 |
5.274 ± 0.265 | 5.274 ± 1.069 |
3.164 ± 0.231 | 6.592 ± 0.329 |
2.373 ± 0.745 | 5.208 ± 0.002 |
5.801 ± 0.533 | 7.383 ± 0.992 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.637 ± 0.04 | 3.955 ± 0.864 |
6.658 ± 0.509 | 4.548 ± 0.103 |
5.933 ± 0.089 | 6.328 ± 0.807 |
7.515 ± 1.698 | 8.833 ± 0.12 |
1.516 ± 0.195 | 2.966 ± 0.102 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
