
Beihai picorna-like virus 120
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
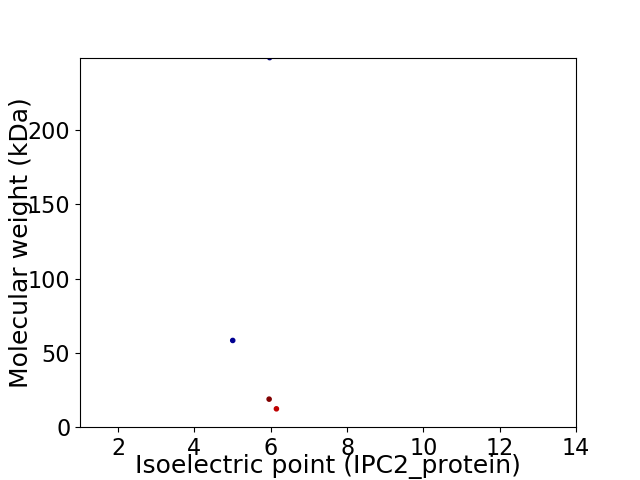
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHP5|A0A1L3KHP5_9VIRU Uncharacterized protein OS=Beihai picorna-like virus 120 OX=1922549 PE=4 SV=1
MM1 pKa = 6.96STRR4 pKa = 11.84AAPATASEE12 pKa = 4.78GVTDD16 pKa = 3.73QHH18 pKa = 6.64PSEE21 pKa = 4.64SGYY24 pKa = 10.7VPNAMGNPAYY34 pKa = 9.4PAIPTRR40 pKa = 11.84TDD42 pKa = 3.56EE43 pKa = 3.85IPEE46 pKa = 4.07RR47 pKa = 11.84VKK49 pKa = 10.82ALNMLDD55 pKa = 3.62PYY57 pKa = 10.68FYY59 pKa = 11.18SQFVLLNSFSWSNQDD74 pKa = 3.5QKK76 pKa = 11.98NKK78 pKa = 9.98ILWSIEE84 pKa = 3.72LKK86 pKa = 10.18PEE88 pKa = 3.86NMHH91 pKa = 7.24ANLAYY96 pKa = 8.44ITAMYY101 pKa = 8.83NAWVGDD107 pKa = 3.83FEE109 pKa = 5.35FMLKK113 pKa = 9.89IAGTGFHH120 pKa = 6.75GGALGVAWIPPNLNVEE136 pKa = 4.4DD137 pKa = 4.48LSGAEE142 pKa = 4.05LTIFDD147 pKa = 4.95YY148 pKa = 11.51LLLDD152 pKa = 4.98PKK154 pKa = 10.52MQDD157 pKa = 3.05LKK159 pKa = 11.4GFAGKK164 pKa = 9.6DD165 pKa = 3.26VKK167 pKa = 11.16NVDD170 pKa = 4.31FHH172 pKa = 6.63WKK174 pKa = 10.19DD175 pKa = 3.33NEE177 pKa = 3.83AAAQFNSDD185 pKa = 3.58FAKK188 pKa = 10.06VASKK192 pKa = 10.9GGYY195 pKa = 9.17LVMFVVLPLITSTTGNNAINLAIFNRR221 pKa = 11.84VAPTFRR227 pKa = 11.84VGQMIPIKK235 pKa = 9.79YY236 pKa = 8.99VKK238 pKa = 9.9PSPALFHH245 pKa = 7.68DD246 pKa = 4.57PFLDD250 pKa = 4.15FSMGVNLRR258 pKa = 11.84NYY260 pKa = 10.08INFGFFKK267 pKa = 10.23QIRR270 pKa = 11.84IEE272 pKa = 4.05PVTIMTTDD280 pKa = 3.09QFRR283 pKa = 11.84DD284 pKa = 3.35LNLSFKK290 pKa = 11.27NMTIVDD296 pKa = 4.28RR297 pKa = 11.84SATSIRR303 pKa = 11.84QVTDD307 pKa = 2.85NGDD310 pKa = 2.9ISFTYY315 pKa = 10.01VRR317 pKa = 11.84SNGVPNPEE325 pKa = 4.88KK326 pKa = 9.81IAKK329 pKa = 10.04GITASQNLVAVEE341 pKa = 4.31WKK343 pKa = 10.69SSANDD348 pKa = 3.51YY349 pKa = 10.35EE350 pKa = 4.8VEE352 pKa = 4.24VANVAGGRR360 pKa = 11.84IGQVHH365 pKa = 7.03DD366 pKa = 3.9AVITYY371 pKa = 8.92GALYY375 pKa = 10.41QSDD378 pKa = 4.27ASWFPNFTNSGGFLTITNGLDD399 pKa = 3.05EE400 pKa = 4.9SLVTFCGQSASNAGFTLPGTVVEE423 pKa = 4.12MLQRR427 pKa = 11.84GMFKK431 pKa = 10.63GIPDD435 pKa = 4.25GQDD438 pKa = 2.72VVYY441 pKa = 10.42DD442 pKa = 3.61IVDD445 pKa = 3.39IPTDD449 pKa = 3.39LVTRR453 pKa = 11.84QIRR456 pKa = 11.84LTEE459 pKa = 3.75IDD461 pKa = 3.36GSVFFTTNKK470 pKa = 8.5TDD472 pKa = 3.3QPIILEE478 pKa = 4.04ASKK481 pKa = 11.19SKK483 pKa = 10.59FVFSEE488 pKa = 4.65FIGRR492 pKa = 11.84EE493 pKa = 4.04TPLTAPAAEE502 pKa = 4.27MKK504 pKa = 10.48RR505 pKa = 11.84NLEE508 pKa = 3.73IVLQRR513 pKa = 11.84DD514 pKa = 3.2RR515 pKa = 11.84LAKK518 pKa = 9.99IEE520 pKa = 3.94QAMAEE525 pKa = 4.25SQMM528 pKa = 3.85
MM1 pKa = 6.96STRR4 pKa = 11.84AAPATASEE12 pKa = 4.78GVTDD16 pKa = 3.73QHH18 pKa = 6.64PSEE21 pKa = 4.64SGYY24 pKa = 10.7VPNAMGNPAYY34 pKa = 9.4PAIPTRR40 pKa = 11.84TDD42 pKa = 3.56EE43 pKa = 3.85IPEE46 pKa = 4.07RR47 pKa = 11.84VKK49 pKa = 10.82ALNMLDD55 pKa = 3.62PYY57 pKa = 10.68FYY59 pKa = 11.18SQFVLLNSFSWSNQDD74 pKa = 3.5QKK76 pKa = 11.98NKK78 pKa = 9.98ILWSIEE84 pKa = 3.72LKK86 pKa = 10.18PEE88 pKa = 3.86NMHH91 pKa = 7.24ANLAYY96 pKa = 8.44ITAMYY101 pKa = 8.83NAWVGDD107 pKa = 3.83FEE109 pKa = 5.35FMLKK113 pKa = 9.89IAGTGFHH120 pKa = 6.75GGALGVAWIPPNLNVEE136 pKa = 4.4DD137 pKa = 4.48LSGAEE142 pKa = 4.05LTIFDD147 pKa = 4.95YY148 pKa = 11.51LLLDD152 pKa = 4.98PKK154 pKa = 10.52MQDD157 pKa = 3.05LKK159 pKa = 11.4GFAGKK164 pKa = 9.6DD165 pKa = 3.26VKK167 pKa = 11.16NVDD170 pKa = 4.31FHH172 pKa = 6.63WKK174 pKa = 10.19DD175 pKa = 3.33NEE177 pKa = 3.83AAAQFNSDD185 pKa = 3.58FAKK188 pKa = 10.06VASKK192 pKa = 10.9GGYY195 pKa = 9.17LVMFVVLPLITSTTGNNAINLAIFNRR221 pKa = 11.84VAPTFRR227 pKa = 11.84VGQMIPIKK235 pKa = 9.79YY236 pKa = 8.99VKK238 pKa = 9.9PSPALFHH245 pKa = 7.68DD246 pKa = 4.57PFLDD250 pKa = 4.15FSMGVNLRR258 pKa = 11.84NYY260 pKa = 10.08INFGFFKK267 pKa = 10.23QIRR270 pKa = 11.84IEE272 pKa = 4.05PVTIMTTDD280 pKa = 3.09QFRR283 pKa = 11.84DD284 pKa = 3.35LNLSFKK290 pKa = 11.27NMTIVDD296 pKa = 4.28RR297 pKa = 11.84SATSIRR303 pKa = 11.84QVTDD307 pKa = 2.85NGDD310 pKa = 2.9ISFTYY315 pKa = 10.01VRR317 pKa = 11.84SNGVPNPEE325 pKa = 4.88KK326 pKa = 9.81IAKK329 pKa = 10.04GITASQNLVAVEE341 pKa = 4.31WKK343 pKa = 10.69SSANDD348 pKa = 3.51YY349 pKa = 10.35EE350 pKa = 4.8VEE352 pKa = 4.24VANVAGGRR360 pKa = 11.84IGQVHH365 pKa = 7.03DD366 pKa = 3.9AVITYY371 pKa = 8.92GALYY375 pKa = 10.41QSDD378 pKa = 4.27ASWFPNFTNSGGFLTITNGLDD399 pKa = 3.05EE400 pKa = 4.9SLVTFCGQSASNAGFTLPGTVVEE423 pKa = 4.12MLQRR427 pKa = 11.84GMFKK431 pKa = 10.63GIPDD435 pKa = 4.25GQDD438 pKa = 2.72VVYY441 pKa = 10.42DD442 pKa = 3.61IVDD445 pKa = 3.39IPTDD449 pKa = 3.39LVTRR453 pKa = 11.84QIRR456 pKa = 11.84LTEE459 pKa = 3.75IDD461 pKa = 3.36GSVFFTTNKK470 pKa = 8.5TDD472 pKa = 3.3QPIILEE478 pKa = 4.04ASKK481 pKa = 11.19SKK483 pKa = 10.59FVFSEE488 pKa = 4.65FIGRR492 pKa = 11.84EE493 pKa = 4.04TPLTAPAAEE502 pKa = 4.27MKK504 pKa = 10.48RR505 pKa = 11.84NLEE508 pKa = 3.73IVLQRR513 pKa = 11.84DD514 pKa = 3.2RR515 pKa = 11.84LAKK518 pKa = 9.99IEE520 pKa = 3.94QAMAEE525 pKa = 4.25SQMM528 pKa = 3.85
Molecular weight: 58.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHP1|A0A1L3KHP1_9VIRU Calici_coat domain-containing protein OS=Beihai picorna-like virus 120 OX=1922549 PE=4 SV=1
MM1 pKa = 7.32AAVAADD7 pKa = 3.86AAAGEE12 pKa = 4.3ASAGAEE18 pKa = 4.19GGSSGISQGASNIAGGLMKK37 pKa = 10.57GAFGLIGQRR46 pKa = 11.84VGADD50 pKa = 3.15LQLRR54 pKa = 11.84NFKK57 pKa = 11.18AEE59 pKa = 3.81GDD61 pKa = 3.82YY62 pKa = 11.0LQNSFQSAGLPGVLAFANGGSSLPIMNQRR91 pKa = 11.84IGQTGLNTLQINSFTPYY108 pKa = 10.16TGTLAQNMFHH118 pKa = 6.97VGKK121 pKa = 10.33LWW123 pKa = 3.28
MM1 pKa = 7.32AAVAADD7 pKa = 3.86AAAGEE12 pKa = 4.3ASAGAEE18 pKa = 4.19GGSSGISQGASNIAGGLMKK37 pKa = 10.57GAFGLIGQRR46 pKa = 11.84VGADD50 pKa = 3.15LQLRR54 pKa = 11.84NFKK57 pKa = 11.18AEE59 pKa = 3.81GDD61 pKa = 3.82YY62 pKa = 11.0LQNSFQSAGLPGVLAFANGGSSLPIMNQRR91 pKa = 11.84IGQTGLNTLQINSFTPYY108 pKa = 10.16TGTLAQNMFHH118 pKa = 6.97VGKK121 pKa = 10.33LWW123 pKa = 3.28
Molecular weight: 12.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3001 |
123 |
2178 |
750.3 |
84.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.864 ± 1.444 | 1.766 ± 0.681 |
6.165 ± 0.564 | 6.265 ± 1.163 |
4.798 ± 0.572 | 6.331 ± 1.611 |
2.299 ± 0.575 | 5.398 ± 0.706 |
6.564 ± 1.014 | 8.031 ± 0.413 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.132 ± 0.099 | 4.432 ± 0.86 |
4.765 ± 0.313 | 4.532 ± 0.334 |
4.732 ± 0.835 | 6.165 ± 1.84 |
5.831 ± 0.338 | 7.164 ± 0.478 |
1.433 ± 0.093 | 3.332 ± 0.324 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
