
Balneola sp.
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Balneolaeota; Balneolia; Balneolales; Balneolaceae; Balneola; unclassified Balneola
Average proteome isoelectric point is 5.66
Get precalculated fractions of proteins
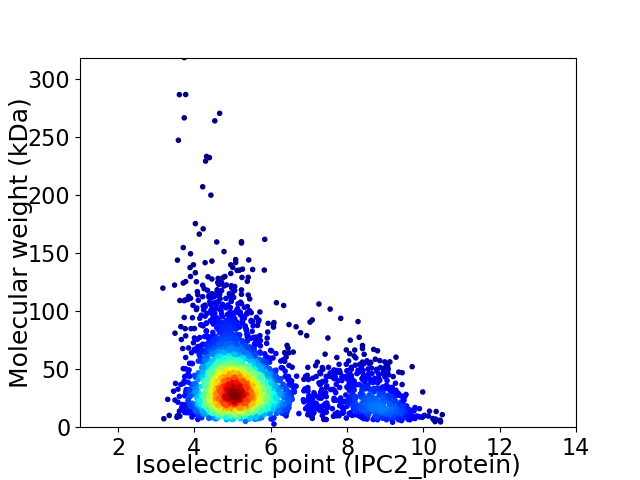
Virtual 2D-PAGE plot for 3130 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
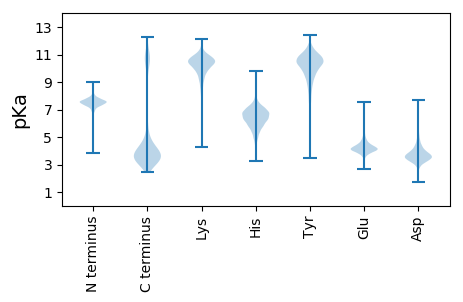
Protein with the lowest isoelectric point:
>tr|A0A3M8G177|A0A3M8G177_9BACT Four helix bundle protein OS=Balneola sp. OX=2024824 GN=ED557_09380 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 10.28KK3 pKa = 10.57LILIPVLIIIAFISCKK19 pKa = 10.32DD20 pKa = 3.53NPAGISGEE28 pKa = 4.11YY29 pKa = 10.85GEE31 pKa = 4.45VNSWIRR37 pKa = 11.84TQMDD41 pKa = 4.61FYY43 pKa = 11.02YY44 pKa = 10.54FWDD47 pKa = 3.53QYY49 pKa = 11.78VPDD52 pKa = 4.21EE53 pKa = 4.68ADD55 pKa = 3.19GTVAPEE61 pKa = 3.74VFYY64 pKa = 11.13EE65 pKa = 5.0SIQEE69 pKa = 4.05PNDD72 pKa = 3.44RR73 pKa = 11.84FSFISDD79 pKa = 3.45DD80 pKa = 3.87AQALLDD86 pKa = 4.42DD87 pKa = 4.85LSGNSVSAGYY97 pKa = 10.24SPAFGRR103 pKa = 11.84ISSTDD108 pKa = 3.03EE109 pKa = 3.59VFIVVEE115 pKa = 4.86FVYY118 pKa = 10.32PGTPAADD125 pKa = 3.33GGMEE129 pKa = 4.28RR130 pKa = 11.84GDD132 pKa = 4.14IILGINGIRR141 pKa = 11.84LDD143 pKa = 3.74TLNYY147 pKa = 10.28LDD149 pKa = 4.9LFYY152 pKa = 10.92TEE154 pKa = 4.95GMSTFTMGEE163 pKa = 3.95ANFSEE168 pKa = 4.53EE169 pKa = 3.94EE170 pKa = 3.52QRR172 pKa = 11.84YY173 pKa = 9.18ILTEE177 pKa = 3.76TGEE180 pKa = 4.44TITADD185 pKa = 3.15VGQIEE190 pKa = 5.32LDD192 pKa = 3.57PVVYY196 pKa = 9.93TSIIDD201 pKa = 3.75TSAHH205 pKa = 6.09KK206 pKa = 9.84IGYY209 pKa = 7.7MFYY212 pKa = 10.87SRR214 pKa = 11.84FVDD217 pKa = 3.47GDD219 pKa = 3.28EE220 pKa = 5.38DD221 pKa = 4.08IFVTSLINTLTDD233 pKa = 3.74FNNQGVTEE241 pKa = 4.16LVVDD245 pKa = 4.13LRR247 pKa = 11.84YY248 pKa = 10.58NPGGQVSVATEE259 pKa = 3.9FANALVPSINAQNEE273 pKa = 4.52DD274 pKa = 2.78IFIQYY279 pKa = 9.41EE280 pKa = 4.18YY281 pKa = 11.37NEE283 pKa = 4.85EE284 pKa = 3.96YY285 pKa = 10.73TNALIDD291 pKa = 3.8QQGLDD296 pKa = 4.08SPNLSTRR303 pKa = 11.84FSEE306 pKa = 4.9GDD308 pKa = 3.32VNLNLDD314 pKa = 3.32KK315 pKa = 11.26VYY317 pKa = 10.84FLVTGASASASEE329 pKa = 4.78LIVTGLEE336 pKa = 4.25PYY338 pKa = 9.67MDD340 pKa = 4.26VYY342 pKa = 11.07TIGTPTVGKK351 pKa = 10.08FYY353 pKa = 10.96GAFVLADD360 pKa = 3.74DD361 pKa = 4.12VSNYY365 pKa = 10.67AIVPVSFKK373 pKa = 10.9YY374 pKa = 10.87LNADD378 pKa = 3.53GFTDD382 pKa = 4.81FASGLDD388 pKa = 3.56PDD390 pKa = 4.95FSTFEE395 pKa = 4.3SLFATVPIGDD405 pKa = 4.44LNDD408 pKa = 4.18PLFSTAIEE416 pKa = 4.56HH417 pKa = 6.15IVNGSVVTKK426 pKa = 8.78PLPFTRR432 pKa = 11.84EE433 pKa = 4.43FIPLRR438 pKa = 11.84DD439 pKa = 4.31PIEE442 pKa = 4.24LKK444 pKa = 10.77NGNVLLDD451 pKa = 3.92FPEE454 pKa = 4.26EE455 pKa = 4.02FF456 pKa = 4.27
MM1 pKa = 7.64KK2 pKa = 10.28KK3 pKa = 10.57LILIPVLIIIAFISCKK19 pKa = 10.32DD20 pKa = 3.53NPAGISGEE28 pKa = 4.11YY29 pKa = 10.85GEE31 pKa = 4.45VNSWIRR37 pKa = 11.84TQMDD41 pKa = 4.61FYY43 pKa = 11.02YY44 pKa = 10.54FWDD47 pKa = 3.53QYY49 pKa = 11.78VPDD52 pKa = 4.21EE53 pKa = 4.68ADD55 pKa = 3.19GTVAPEE61 pKa = 3.74VFYY64 pKa = 11.13EE65 pKa = 5.0SIQEE69 pKa = 4.05PNDD72 pKa = 3.44RR73 pKa = 11.84FSFISDD79 pKa = 3.45DD80 pKa = 3.87AQALLDD86 pKa = 4.42DD87 pKa = 4.85LSGNSVSAGYY97 pKa = 10.24SPAFGRR103 pKa = 11.84ISSTDD108 pKa = 3.03EE109 pKa = 3.59VFIVVEE115 pKa = 4.86FVYY118 pKa = 10.32PGTPAADD125 pKa = 3.33GGMEE129 pKa = 4.28RR130 pKa = 11.84GDD132 pKa = 4.14IILGINGIRR141 pKa = 11.84LDD143 pKa = 3.74TLNYY147 pKa = 10.28LDD149 pKa = 4.9LFYY152 pKa = 10.92TEE154 pKa = 4.95GMSTFTMGEE163 pKa = 3.95ANFSEE168 pKa = 4.53EE169 pKa = 3.94EE170 pKa = 3.52QRR172 pKa = 11.84YY173 pKa = 9.18ILTEE177 pKa = 3.76TGEE180 pKa = 4.44TITADD185 pKa = 3.15VGQIEE190 pKa = 5.32LDD192 pKa = 3.57PVVYY196 pKa = 9.93TSIIDD201 pKa = 3.75TSAHH205 pKa = 6.09KK206 pKa = 9.84IGYY209 pKa = 7.7MFYY212 pKa = 10.87SRR214 pKa = 11.84FVDD217 pKa = 3.47GDD219 pKa = 3.28EE220 pKa = 5.38DD221 pKa = 4.08IFVTSLINTLTDD233 pKa = 3.74FNNQGVTEE241 pKa = 4.16LVVDD245 pKa = 4.13LRR247 pKa = 11.84YY248 pKa = 10.58NPGGQVSVATEE259 pKa = 3.9FANALVPSINAQNEE273 pKa = 4.52DD274 pKa = 2.78IFIQYY279 pKa = 9.41EE280 pKa = 4.18YY281 pKa = 11.37NEE283 pKa = 4.85EE284 pKa = 3.96YY285 pKa = 10.73TNALIDD291 pKa = 3.8QQGLDD296 pKa = 4.08SPNLSTRR303 pKa = 11.84FSEE306 pKa = 4.9GDD308 pKa = 3.32VNLNLDD314 pKa = 3.32KK315 pKa = 11.26VYY317 pKa = 10.84FLVTGASASASEE329 pKa = 4.78LIVTGLEE336 pKa = 4.25PYY338 pKa = 9.67MDD340 pKa = 4.26VYY342 pKa = 11.07TIGTPTVGKK351 pKa = 10.08FYY353 pKa = 10.96GAFVLADD360 pKa = 3.74DD361 pKa = 4.12VSNYY365 pKa = 10.67AIVPVSFKK373 pKa = 10.9YY374 pKa = 10.87LNADD378 pKa = 3.53GFTDD382 pKa = 4.81FASGLDD388 pKa = 3.56PDD390 pKa = 4.95FSTFEE395 pKa = 4.3SLFATVPIGDD405 pKa = 4.44LNDD408 pKa = 4.18PLFSTAIEE416 pKa = 4.56HH417 pKa = 6.15IVNGSVVTKK426 pKa = 8.78PLPFTRR432 pKa = 11.84EE433 pKa = 4.43FIPLRR438 pKa = 11.84DD439 pKa = 4.31PIEE442 pKa = 4.24LKK444 pKa = 10.77NGNVLLDD451 pKa = 3.92FPEE454 pKa = 4.26EE455 pKa = 4.02FF456 pKa = 4.27
Molecular weight: 50.41 kDa
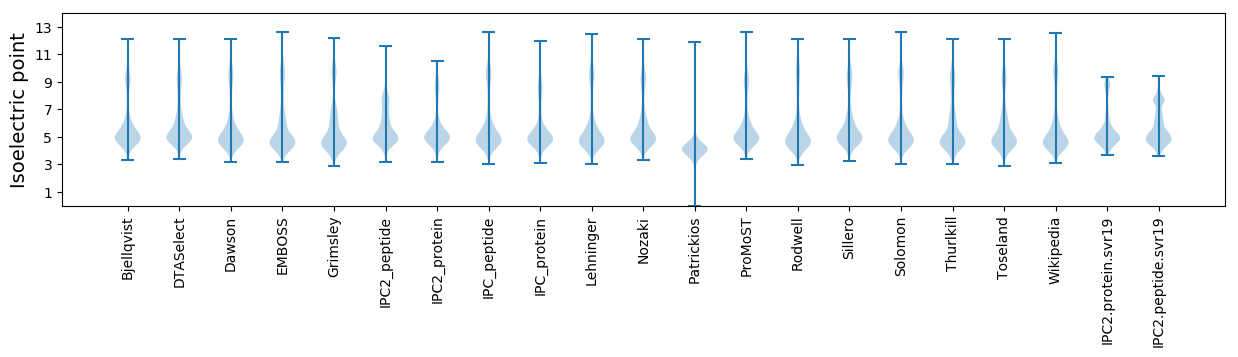
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M8FQ57|A0A3M8FQ57_9BACT Sensor histidine kinase OS=Balneola sp. OX=2024824 GN=ED557_14720 PE=4 SV=1
MM1 pKa = 7.57AHH3 pKa = 7.61PKK5 pKa = 10.33RR6 pKa = 11.84KK7 pKa = 7.54TSKK10 pKa = 9.88ARR12 pKa = 11.84RR13 pKa = 11.84DD14 pKa = 3.43KK15 pKa = 10.9RR16 pKa = 11.84RR17 pKa = 11.84SHH19 pKa = 6.51HH20 pKa = 6.92AIADD24 pKa = 3.6VTLAKK29 pKa = 10.41CSNCGTLHH37 pKa = 7.19RR38 pKa = 11.84YY39 pKa = 7.94HH40 pKa = 6.85HH41 pKa = 7.03ACPEE45 pKa = 3.84CGYY48 pKa = 10.54YY49 pKa = 10.03RR50 pKa = 11.84GRR52 pKa = 11.84QVISVANN59 pKa = 3.74
MM1 pKa = 7.57AHH3 pKa = 7.61PKK5 pKa = 10.33RR6 pKa = 11.84KK7 pKa = 7.54TSKK10 pKa = 9.88ARR12 pKa = 11.84RR13 pKa = 11.84DD14 pKa = 3.43KK15 pKa = 10.9RR16 pKa = 11.84RR17 pKa = 11.84SHH19 pKa = 6.51HH20 pKa = 6.92AIADD24 pKa = 3.6VTLAKK29 pKa = 10.41CSNCGTLHH37 pKa = 7.19RR38 pKa = 11.84YY39 pKa = 7.94HH40 pKa = 6.85HH41 pKa = 7.03ACPEE45 pKa = 3.84CGYY48 pKa = 10.54YY49 pKa = 10.03RR50 pKa = 11.84GRR52 pKa = 11.84QVISVANN59 pKa = 3.74
Molecular weight: 6.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1114347 |
22 |
3034 |
356.0 |
39.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.472 ± 0.038 | 0.575 ± 0.011 |
5.889 ± 0.041 | 7.478 ± 0.049 |
5.234 ± 0.037 | 7.108 ± 0.041 |
1.78 ± 0.024 | 7.465 ± 0.04 |
5.524 ± 0.047 | 9.396 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.157 ± 0.02 | 5.071 ± 0.036 |
3.772 ± 0.024 | 3.415 ± 0.024 |
4.205 ± 0.029 | 7.339 ± 0.043 |
5.69 ± 0.049 | 6.431 ± 0.032 |
1.223 ± 0.017 | 3.774 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
