
alpha proteobacterium BAL199
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; unclassified Alphaproteobacteria
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
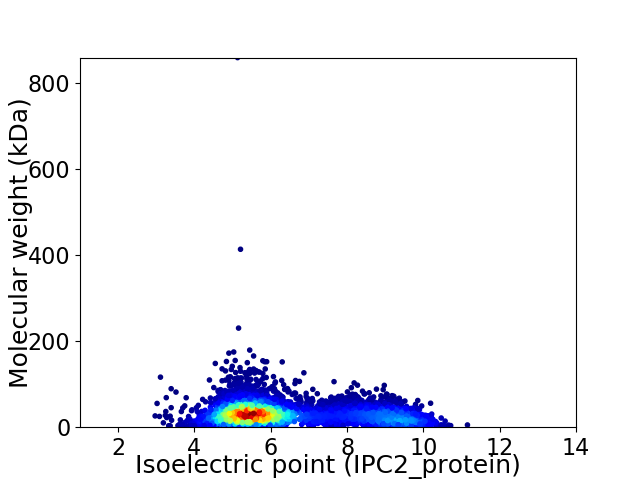
Virtual 2D-PAGE plot for 5881 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8TNX4|A8TNX4_9PROT DUF126 domain-containing protein OS=alpha proteobacterium BAL199 OX=331869 GN=BAL199_12591 PE=4 SV=1
MM1 pKa = 7.27TVLGTDD7 pKa = 4.01DD8 pKa = 3.77RR9 pKa = 11.84VQVADD14 pKa = 4.01TTAFPYY20 pKa = 10.42SAIVQVRR27 pKa = 11.84VDD29 pKa = 3.94FDD31 pKa = 4.47GDD33 pKa = 3.3GSYY36 pKa = 11.15DD37 pKa = 3.26GWGTGAMISPNDD49 pKa = 3.66VLTAGHH55 pKa = 6.05VLWDD59 pKa = 3.65SQYY62 pKa = 11.47GYY64 pKa = 11.08AKK66 pKa = 10.2NIQVIPARR74 pKa = 11.84DD75 pKa = 3.58GGSTPFGTAEE85 pKa = 3.78GTAWYY90 pKa = 10.4VPDD93 pKa = 5.05AYY95 pKa = 9.78ISTGGTFAYY104 pKa = 10.41DD105 pKa = 2.69IGVINLGTSIGSSTGTLALQATSGPDD131 pKa = 3.33LVGTTVTTAGYY142 pKa = 9.53PGDD145 pKa = 3.87LSSNGSVMITTSGEE159 pKa = 3.66IDD161 pKa = 3.67GSIGDD166 pKa = 3.44NRR168 pKa = 11.84VYY170 pKa = 11.45YY171 pKa = 10.8NGTLDD176 pKa = 3.67SYY178 pKa = 11.26GGQSGSPLWVTVNGQTTIVGVHH200 pKa = 4.84TAGGSFYY207 pKa = 11.02NAGTALTSDD216 pKa = 5.07FYY218 pKa = 11.91SLIDD222 pKa = 3.34GWTGGDD228 pKa = 3.05INGSVVVVTSSTSGTASADD247 pKa = 3.68VITGSTGSDD256 pKa = 3.11TVSGYY261 pKa = 11.23AGDD264 pKa = 3.72DD265 pKa = 4.19TIWGEE270 pKa = 4.05SGNDD274 pKa = 3.24VIYY277 pKa = 10.69GNQGADD283 pKa = 4.01LISGQDD289 pKa = 3.5GNDD292 pKa = 3.75TIFGGQNQGPAGSDD306 pKa = 3.56GIARR310 pKa = 11.84NGTDD314 pKa = 3.96TIQGGDD320 pKa = 3.72GADD323 pKa = 3.14VLYY326 pKa = 11.29GNMGGDD332 pKa = 4.16LVDD335 pKa = 4.74GDD337 pKa = 4.39SGDD340 pKa = 4.65DD341 pKa = 3.33ILYY344 pKa = 10.59GGQDD348 pKa = 3.18SDD350 pKa = 3.95TLRR353 pKa = 11.84GGDD356 pKa = 3.3GADD359 pKa = 2.46RR360 pKa = 11.84VFGNLHH366 pKa = 6.79NDD368 pKa = 3.45TLSGGGGNDD377 pKa = 3.29TLLGGDD383 pKa = 4.29GNDD386 pKa = 3.92LLSGGSGDD394 pKa = 4.58DD395 pKa = 4.53LLYY398 pKa = 11.08GDD400 pKa = 4.83NLFSGVVTLTRR411 pKa = 11.84RR412 pKa = 11.84VSKK415 pKa = 9.71LACRR419 pKa = 11.84SLIKK423 pKa = 10.38RR424 pKa = 11.84HH425 pKa = 5.24VPRR428 pKa = 11.84ISKK431 pKa = 10.06LRR433 pKa = 11.84TLRR436 pKa = 11.84ADD438 pKa = 3.27GSEE441 pKa = 3.84WW442 pKa = 3.2
MM1 pKa = 7.27TVLGTDD7 pKa = 4.01DD8 pKa = 3.77RR9 pKa = 11.84VQVADD14 pKa = 4.01TTAFPYY20 pKa = 10.42SAIVQVRR27 pKa = 11.84VDD29 pKa = 3.94FDD31 pKa = 4.47GDD33 pKa = 3.3GSYY36 pKa = 11.15DD37 pKa = 3.26GWGTGAMISPNDD49 pKa = 3.66VLTAGHH55 pKa = 6.05VLWDD59 pKa = 3.65SQYY62 pKa = 11.47GYY64 pKa = 11.08AKK66 pKa = 10.2NIQVIPARR74 pKa = 11.84DD75 pKa = 3.58GGSTPFGTAEE85 pKa = 3.78GTAWYY90 pKa = 10.4VPDD93 pKa = 5.05AYY95 pKa = 9.78ISTGGTFAYY104 pKa = 10.41DD105 pKa = 2.69IGVINLGTSIGSSTGTLALQATSGPDD131 pKa = 3.33LVGTTVTTAGYY142 pKa = 9.53PGDD145 pKa = 3.87LSSNGSVMITTSGEE159 pKa = 3.66IDD161 pKa = 3.67GSIGDD166 pKa = 3.44NRR168 pKa = 11.84VYY170 pKa = 11.45YY171 pKa = 10.8NGTLDD176 pKa = 3.67SYY178 pKa = 11.26GGQSGSPLWVTVNGQTTIVGVHH200 pKa = 4.84TAGGSFYY207 pKa = 11.02NAGTALTSDD216 pKa = 5.07FYY218 pKa = 11.91SLIDD222 pKa = 3.34GWTGGDD228 pKa = 3.05INGSVVVVTSSTSGTASADD247 pKa = 3.68VITGSTGSDD256 pKa = 3.11TVSGYY261 pKa = 11.23AGDD264 pKa = 3.72DD265 pKa = 4.19TIWGEE270 pKa = 4.05SGNDD274 pKa = 3.24VIYY277 pKa = 10.69GNQGADD283 pKa = 4.01LISGQDD289 pKa = 3.5GNDD292 pKa = 3.75TIFGGQNQGPAGSDD306 pKa = 3.56GIARR310 pKa = 11.84NGTDD314 pKa = 3.96TIQGGDD320 pKa = 3.72GADD323 pKa = 3.14VLYY326 pKa = 11.29GNMGGDD332 pKa = 4.16LVDD335 pKa = 4.74GDD337 pKa = 4.39SGDD340 pKa = 4.65DD341 pKa = 3.33ILYY344 pKa = 10.59GGQDD348 pKa = 3.18SDD350 pKa = 3.95TLRR353 pKa = 11.84GGDD356 pKa = 3.3GADD359 pKa = 2.46RR360 pKa = 11.84VFGNLHH366 pKa = 6.79NDD368 pKa = 3.45TLSGGGGNDD377 pKa = 3.29TLLGGDD383 pKa = 4.29GNDD386 pKa = 3.92LLSGGSGDD394 pKa = 4.58DD395 pKa = 4.53LLYY398 pKa = 11.08GDD400 pKa = 4.83NLFSGVVTLTRR411 pKa = 11.84RR412 pKa = 11.84VSKK415 pKa = 9.71LACRR419 pKa = 11.84SLIKK423 pKa = 10.38RR424 pKa = 11.84HH425 pKa = 5.24VPRR428 pKa = 11.84ISKK431 pKa = 10.06LRR433 pKa = 11.84TLRR436 pKa = 11.84ADD438 pKa = 3.27GSEE441 pKa = 3.84WW442 pKa = 3.2
Molecular weight: 44.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8TUE9|A8TUE9_9PROT Probable membrane transporter protein OS=alpha proteobacterium BAL199 OX=331869 GN=BAL199_15107 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.1QPSVLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84RR29 pKa = 11.84VIATRR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.1QPSVLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84RR29 pKa = 11.84VIATRR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1761557 |
13 |
8056 |
299.5 |
32.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.603 ± 0.047 | 0.877 ± 0.01 |
6.199 ± 0.028 | 5.403 ± 0.026 |
3.469 ± 0.02 | 8.741 ± 0.036 |
2.117 ± 0.014 | 4.862 ± 0.021 |
2.693 ± 0.027 | 10.091 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.429 ± 0.016 | 2.299 ± 0.017 |
5.375 ± 0.025 | 2.815 ± 0.018 |
7.767 ± 0.035 | 5.159 ± 0.023 |
5.506 ± 0.019 | 8.019 ± 0.029 |
1.43 ± 0.014 | 2.149 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
