
Circovirus-like genome DHCV-6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.57
Get precalculated fractions of proteins
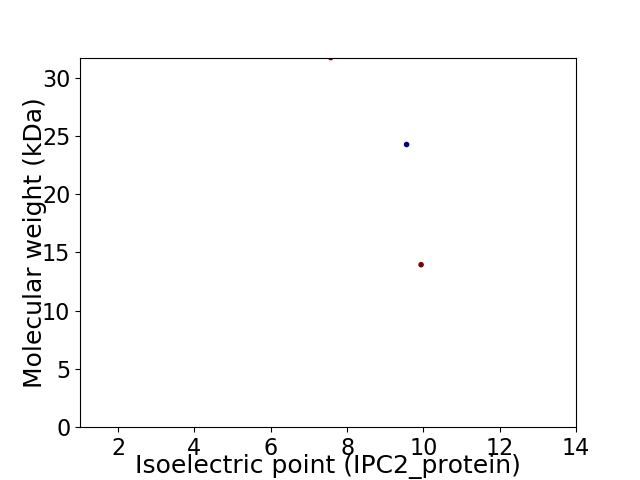
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
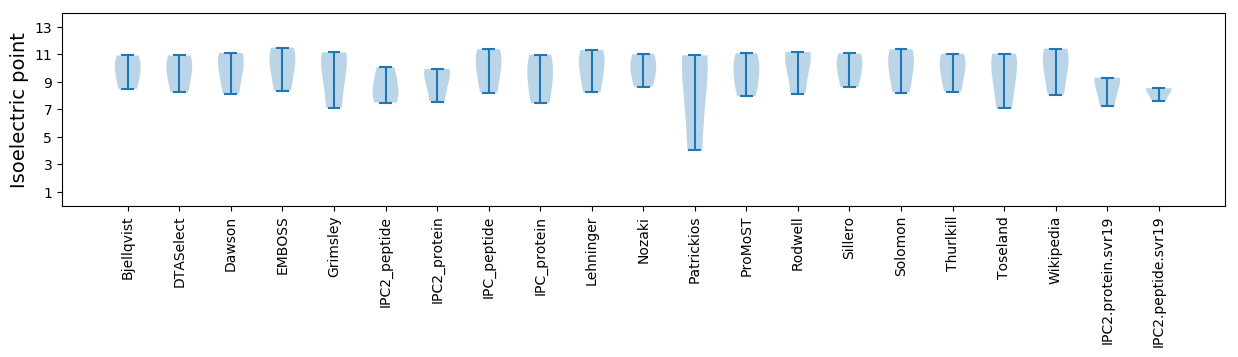
Protein with the lowest isoelectric point:
>tr|A0A190WHH1|A0A190WHH1_9CIRC Uncharacterized protein OS=Circovirus-like genome DHCV-6 OX=1788455 PE=4 SV=1
MM1 pKa = 7.27SRR3 pKa = 11.84HH4 pKa = 5.85RR5 pKa = 11.84NYY7 pKa = 10.67CFTYY11 pKa = 9.97NNYY14 pKa = 9.96PNTEE18 pKa = 3.73LVDD21 pKa = 3.85NVNCKK26 pKa = 10.3YY27 pKa = 10.0IAYY30 pKa = 8.56AHH32 pKa = 6.21EE33 pKa = 4.23VAPTTGTRR41 pKa = 11.84HH42 pKa = 4.78LQGYY46 pKa = 8.95IAFHH50 pKa = 4.88STKK53 pKa = 9.54TLAAARR59 pKa = 11.84RR60 pKa = 11.84LLPGCHH66 pKa = 6.67LSAMAGSISQNDD78 pKa = 4.27AYY80 pKa = 10.66CSKK83 pKa = 10.74SGTLIEE89 pKa = 4.94RR90 pKa = 11.84GEE92 pKa = 4.27KK93 pKa = 9.45PASNDD98 pKa = 3.21DD99 pKa = 3.2KK100 pKa = 11.69GRR102 pKa = 11.84AEE104 pKa = 3.88KK105 pKa = 10.78LRR107 pKa = 11.84WQRR110 pKa = 11.84ARR112 pKa = 11.84DD113 pKa = 3.45LAKK116 pKa = 10.64AGNLDD121 pKa = 4.47EE122 pKa = 5.82IDD124 pKa = 3.18ADD126 pKa = 3.63IYY128 pKa = 10.04IRR130 pKa = 11.84CYY132 pKa = 9.74STLKK136 pKa = 10.68SISKK140 pKa = 10.24DD141 pKa = 3.21HH142 pKa = 6.55MIKK145 pKa = 10.55PEE147 pKa = 4.14PVDD150 pKa = 4.42VKK152 pKa = 10.49CFWIHH157 pKa = 6.23GATGTGKK164 pKa = 10.07SYY166 pKa = 10.77CVEE169 pKa = 3.89TTYY172 pKa = 11.04PDD174 pKa = 3.94CYY176 pKa = 9.91KK177 pKa = 11.1KK178 pKa = 11.3SMDD181 pKa = 3.7DD182 pKa = 3.91LKK184 pKa = 11.14WFDD187 pKa = 4.55GYY189 pKa = 11.55DD190 pKa = 3.7NQDD193 pKa = 2.79VVYY196 pKa = 10.96LEE198 pKa = 5.4DD199 pKa = 4.05FDD201 pKa = 5.32VYY203 pKa = 9.93QIKK206 pKa = 9.79WGGLMKK212 pKa = 10.59RR213 pKa = 11.84LADD216 pKa = 4.28RR217 pKa = 11.84WPMQASIKK225 pKa = 10.8GSMKK229 pKa = 10.25YY230 pKa = 9.82IRR232 pKa = 11.84PKK234 pKa = 10.36IVIVTSNYY242 pKa = 8.38TPNEE246 pKa = 3.36IWMDD250 pKa = 3.74PQTVEE255 pKa = 3.56PLLRR259 pKa = 11.84RR260 pKa = 11.84FKK262 pKa = 10.95VIEE265 pKa = 4.26KK266 pKa = 9.97VSQEE270 pKa = 3.91QVVDD274 pKa = 4.49FTT276 pKa = 4.81
MM1 pKa = 7.27SRR3 pKa = 11.84HH4 pKa = 5.85RR5 pKa = 11.84NYY7 pKa = 10.67CFTYY11 pKa = 9.97NNYY14 pKa = 9.96PNTEE18 pKa = 3.73LVDD21 pKa = 3.85NVNCKK26 pKa = 10.3YY27 pKa = 10.0IAYY30 pKa = 8.56AHH32 pKa = 6.21EE33 pKa = 4.23VAPTTGTRR41 pKa = 11.84HH42 pKa = 4.78LQGYY46 pKa = 8.95IAFHH50 pKa = 4.88STKK53 pKa = 9.54TLAAARR59 pKa = 11.84RR60 pKa = 11.84LLPGCHH66 pKa = 6.67LSAMAGSISQNDD78 pKa = 4.27AYY80 pKa = 10.66CSKK83 pKa = 10.74SGTLIEE89 pKa = 4.94RR90 pKa = 11.84GEE92 pKa = 4.27KK93 pKa = 9.45PASNDD98 pKa = 3.21DD99 pKa = 3.2KK100 pKa = 11.69GRR102 pKa = 11.84AEE104 pKa = 3.88KK105 pKa = 10.78LRR107 pKa = 11.84WQRR110 pKa = 11.84ARR112 pKa = 11.84DD113 pKa = 3.45LAKK116 pKa = 10.64AGNLDD121 pKa = 4.47EE122 pKa = 5.82IDD124 pKa = 3.18ADD126 pKa = 3.63IYY128 pKa = 10.04IRR130 pKa = 11.84CYY132 pKa = 9.74STLKK136 pKa = 10.68SISKK140 pKa = 10.24DD141 pKa = 3.21HH142 pKa = 6.55MIKK145 pKa = 10.55PEE147 pKa = 4.14PVDD150 pKa = 4.42VKK152 pKa = 10.49CFWIHH157 pKa = 6.23GATGTGKK164 pKa = 10.07SYY166 pKa = 10.77CVEE169 pKa = 3.89TTYY172 pKa = 11.04PDD174 pKa = 3.94CYY176 pKa = 9.91KK177 pKa = 11.1KK178 pKa = 11.3SMDD181 pKa = 3.7DD182 pKa = 3.91LKK184 pKa = 11.14WFDD187 pKa = 4.55GYY189 pKa = 11.55DD190 pKa = 3.7NQDD193 pKa = 2.79VVYY196 pKa = 10.96LEE198 pKa = 5.4DD199 pKa = 4.05FDD201 pKa = 5.32VYY203 pKa = 9.93QIKK206 pKa = 9.79WGGLMKK212 pKa = 10.59RR213 pKa = 11.84LADD216 pKa = 4.28RR217 pKa = 11.84WPMQASIKK225 pKa = 10.8GSMKK229 pKa = 10.25YY230 pKa = 9.82IRR232 pKa = 11.84PKK234 pKa = 10.36IVIVTSNYY242 pKa = 8.38TPNEE246 pKa = 3.36IWMDD250 pKa = 3.74PQTVEE255 pKa = 3.56PLLRR259 pKa = 11.84RR260 pKa = 11.84FKK262 pKa = 10.95VIEE265 pKa = 4.26KK266 pKa = 9.97VSQEE270 pKa = 3.91QVVDD274 pKa = 4.49FTT276 pKa = 4.81
Molecular weight: 31.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A190WHP0|A0A190WHP0_9CIRC Uncharacterized protein OS=Circovirus-like genome DHCV-6 OX=1788455 PE=4 SV=1
MM1 pKa = 7.67LIASISLMLMRR12 pKa = 11.84LLQQLAPDD20 pKa = 3.88ISRR23 pKa = 11.84VILLSIQQKK32 pKa = 10.2LLQLLGDD39 pKa = 4.43FYY41 pKa = 11.65RR42 pKa = 11.84DD43 pKa = 3.98AISQLWRR50 pKa = 11.84DD51 pKa = 3.96LYY53 pKa = 10.35PRR55 pKa = 11.84TMRR58 pKa = 11.84TARR61 pKa = 11.84KK62 pKa = 9.02VARR65 pKa = 11.84LLNGGRR71 pKa = 11.84NLLPMTIRR79 pKa = 11.84DD80 pKa = 3.63VPRR83 pKa = 11.84NSDD86 pKa = 3.14GSALEE91 pKa = 3.99ILLKK95 pKa = 10.8LVTWTKK101 pKa = 11.19LMLIFISVAILLLNQYY117 pKa = 10.73LKK119 pKa = 10.54III121 pKa = 4.2
MM1 pKa = 7.67LIASISLMLMRR12 pKa = 11.84LLQQLAPDD20 pKa = 3.88ISRR23 pKa = 11.84VILLSIQQKK32 pKa = 10.2LLQLLGDD39 pKa = 4.43FYY41 pKa = 11.65RR42 pKa = 11.84DD43 pKa = 3.98AISQLWRR50 pKa = 11.84DD51 pKa = 3.96LYY53 pKa = 10.35PRR55 pKa = 11.84TMRR58 pKa = 11.84TARR61 pKa = 11.84KK62 pKa = 9.02VARR65 pKa = 11.84LLNGGRR71 pKa = 11.84NLLPMTIRR79 pKa = 11.84DD80 pKa = 3.63VPRR83 pKa = 11.84NSDD86 pKa = 3.14GSALEE91 pKa = 3.99ILLKK95 pKa = 10.8LVTWTKK101 pKa = 11.19LMLIFISVAILLLNQYY117 pKa = 10.73LKK119 pKa = 10.54III121 pKa = 4.2
Molecular weight: 13.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
619 |
121 |
276 |
206.3 |
23.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.947 ± 0.399 | 1.292 ± 0.83 |
5.493 ± 1.164 | 2.908 ± 0.95 |
3.231 ± 0.792 | 5.977 ± 1.061 |
1.454 ± 0.588 | 6.947 ± 1.107 |
7.108 ± 0.864 | 10.662 ± 3.689 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.231 ± 0.506 | 4.362 ± 0.35 |
3.393 ± 0.545 | 4.685 ± 0.736 |
5.654 ± 1.167 | 7.431 ± 0.899 |
5.816 ± 0.491 | 7.108 ± 1.494 |
1.292 ± 0.579 | 5.008 ± 0.787 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
