
Rhodococcus tukisamuensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Nocardiaceae; Rhodococcus
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
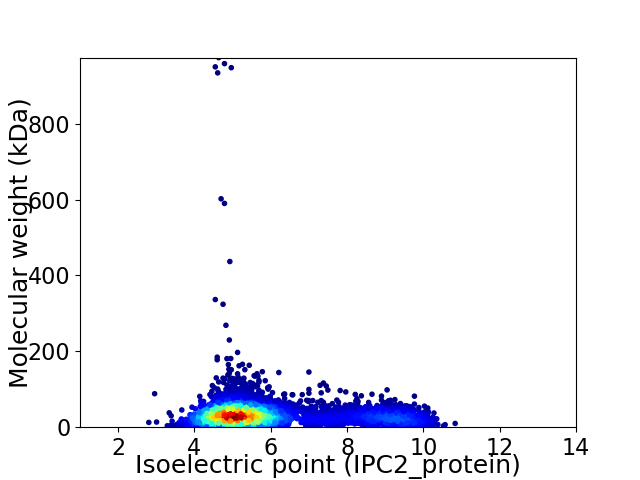
Virtual 2D-PAGE plot for 4987 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G7BGF2|A0A1G7BGF2_9NOCA Uncharacterized protein OS=Rhodococcus tukisamuensis OX=168276 GN=SAMN05444580_11321 PE=4 SV=1
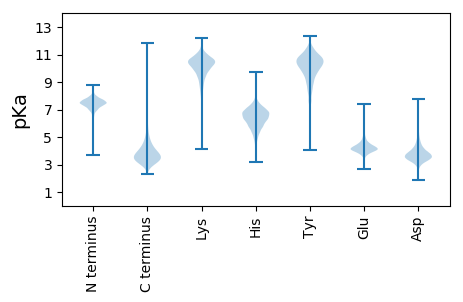
MM1 pKa = 7.62TEE3 pKa = 5.51LDD5 pKa = 3.93SQDD8 pKa = 3.12QLLRR12 pKa = 11.84IDD14 pKa = 4.34GEE16 pKa = 4.19LAIATEE22 pKa = 4.58GHH24 pKa = 6.43DD25 pKa = 3.94QLHH28 pKa = 6.2ALCLRR33 pKa = 11.84TLAEE37 pKa = 4.44LLGSEE42 pKa = 4.71PDD44 pKa = 3.25VDD46 pKa = 3.97EE47 pKa = 6.24DD48 pKa = 4.12GDD50 pKa = 3.85IAIPVHH56 pKa = 6.02GFVVYY61 pKa = 8.04VTVADD66 pKa = 5.23DD67 pKa = 4.55GPQLHH72 pKa = 6.26VWSSILTGLGDD83 pKa = 3.43RR84 pKa = 11.84ARR86 pKa = 11.84AAEE89 pKa = 4.29ALTDD93 pKa = 5.57LALEE97 pKa = 4.29WPRR100 pKa = 11.84LRR102 pKa = 11.84FVFSDD107 pKa = 3.24DD108 pKa = 3.58HH109 pKa = 8.24LLVSTILDD117 pKa = 3.81AEE119 pKa = 4.34PFAPQHH125 pKa = 6.42LINLVDD131 pKa = 3.89EE132 pKa = 4.44MHH134 pKa = 6.87EE135 pKa = 4.22LTHH138 pKa = 7.62DD139 pKa = 4.62LDD141 pKa = 5.67DD142 pKa = 5.03DD143 pKa = 3.94LAARR147 pKa = 11.84FGGTLDD153 pKa = 4.98CDD155 pKa = 4.17TEE157 pKa = 5.0DD158 pKa = 3.66DD159 pKa = 4.71SYY161 pKa = 12.19AGGCGGCGDD170 pKa = 5.71DD171 pKa = 5.44CDD173 pKa = 4.99CDD175 pKa = 5.29CGDD178 pKa = 3.94TDD180 pKa = 5.06EE181 pKa = 4.57GHH183 pKa = 7.52LDD185 pKa = 3.13ITAVNPSKK193 pKa = 11.14
MM1 pKa = 7.62TEE3 pKa = 5.51LDD5 pKa = 3.93SQDD8 pKa = 3.12QLLRR12 pKa = 11.84IDD14 pKa = 4.34GEE16 pKa = 4.19LAIATEE22 pKa = 4.58GHH24 pKa = 6.43DD25 pKa = 3.94QLHH28 pKa = 6.2ALCLRR33 pKa = 11.84TLAEE37 pKa = 4.44LLGSEE42 pKa = 4.71PDD44 pKa = 3.25VDD46 pKa = 3.97EE47 pKa = 6.24DD48 pKa = 4.12GDD50 pKa = 3.85IAIPVHH56 pKa = 6.02GFVVYY61 pKa = 8.04VTVADD66 pKa = 5.23DD67 pKa = 4.55GPQLHH72 pKa = 6.26VWSSILTGLGDD83 pKa = 3.43RR84 pKa = 11.84ARR86 pKa = 11.84AAEE89 pKa = 4.29ALTDD93 pKa = 5.57LALEE97 pKa = 4.29WPRR100 pKa = 11.84LRR102 pKa = 11.84FVFSDD107 pKa = 3.24DD108 pKa = 3.58HH109 pKa = 8.24LLVSTILDD117 pKa = 3.81AEE119 pKa = 4.34PFAPQHH125 pKa = 6.42LINLVDD131 pKa = 3.89EE132 pKa = 4.44MHH134 pKa = 6.87EE135 pKa = 4.22LTHH138 pKa = 7.62DD139 pKa = 4.62LDD141 pKa = 5.67DD142 pKa = 5.03DD143 pKa = 3.94LAARR147 pKa = 11.84FGGTLDD153 pKa = 4.98CDD155 pKa = 4.17TEE157 pKa = 5.0DD158 pKa = 3.66DD159 pKa = 4.71SYY161 pKa = 12.19AGGCGGCGDD170 pKa = 5.71DD171 pKa = 5.44CDD173 pKa = 4.99CDD175 pKa = 5.29CGDD178 pKa = 3.94TDD180 pKa = 5.06EE181 pKa = 4.57GHH183 pKa = 7.52LDD185 pKa = 3.13ITAVNPSKK193 pKa = 11.14
Molecular weight: 20.83 kDa
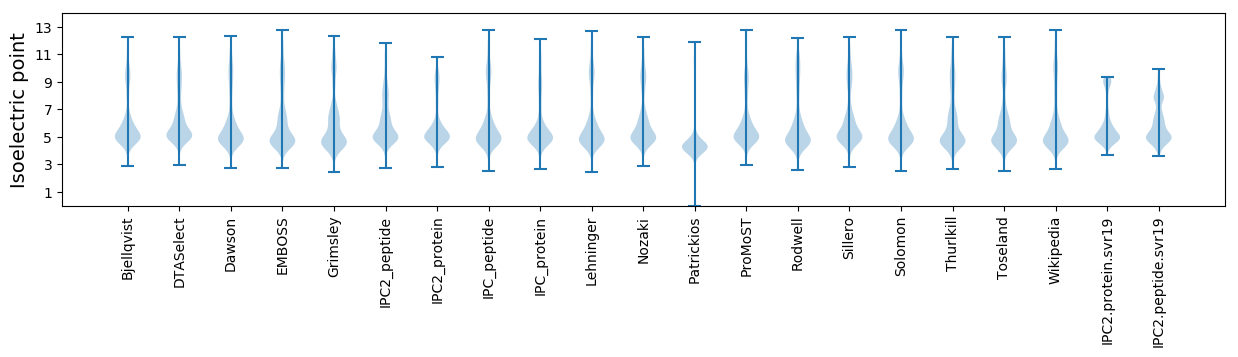
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G7B447|A0A1G7B447_9NOCA L-lactate dehydrogenase complex protein LldG OS=Rhodococcus tukisamuensis OX=168276 GN=SAMN05444580_11263 PE=4 SV=1
MM1 pKa = 8.07DD2 pKa = 4.41PTASTDD8 pKa = 3.29RR9 pKa = 11.84RR10 pKa = 11.84DD11 pKa = 3.39AAVATVTDD19 pKa = 4.67FFAALDD25 pKa = 3.93ALDD28 pKa = 3.75VPRR31 pKa = 11.84AGTFLTPDD39 pKa = 3.56VVWRR43 pKa = 11.84NTTLPTLRR51 pKa = 11.84GLPTVTRR58 pKa = 11.84ALRR61 pKa = 11.84AAARR65 pKa = 11.84PRR67 pKa = 11.84CRR69 pKa = 11.84FEE71 pKa = 4.86AVVHH75 pKa = 6.45HH76 pKa = 6.71IASDD80 pKa = 3.5GDD82 pKa = 3.7AVLVEE87 pKa = 4.33RR88 pKa = 11.84TDD90 pKa = 3.55TLTLGPLRR98 pKa = 11.84STFWVCGTFEE108 pKa = 4.18IRR110 pKa = 11.84DD111 pKa = 3.51GKK113 pKa = 9.33IAVWRR118 pKa = 11.84DD119 pKa = 3.09YY120 pKa = 11.03FSWRR124 pKa = 11.84NVLLGAVTGTLRR136 pKa = 11.84SVRR139 pKa = 11.84GRR141 pKa = 11.84QRR143 pKa = 11.84HH144 pKa = 5.33LPAAPGHH151 pKa = 5.7GRR153 pKa = 11.84STGTIPGAAPATVPPDD169 pKa = 3.71YY170 pKa = 10.3PRR172 pKa = 11.84KK173 pKa = 9.88DD174 pKa = 3.34QVKK177 pKa = 9.87
MM1 pKa = 8.07DD2 pKa = 4.41PTASTDD8 pKa = 3.29RR9 pKa = 11.84RR10 pKa = 11.84DD11 pKa = 3.39AAVATVTDD19 pKa = 4.67FFAALDD25 pKa = 3.93ALDD28 pKa = 3.75VPRR31 pKa = 11.84AGTFLTPDD39 pKa = 3.56VVWRR43 pKa = 11.84NTTLPTLRR51 pKa = 11.84GLPTVTRR58 pKa = 11.84ALRR61 pKa = 11.84AAARR65 pKa = 11.84PRR67 pKa = 11.84CRR69 pKa = 11.84FEE71 pKa = 4.86AVVHH75 pKa = 6.45HH76 pKa = 6.71IASDD80 pKa = 3.5GDD82 pKa = 3.7AVLVEE87 pKa = 4.33RR88 pKa = 11.84TDD90 pKa = 3.55TLTLGPLRR98 pKa = 11.84STFWVCGTFEE108 pKa = 4.18IRR110 pKa = 11.84DD111 pKa = 3.51GKK113 pKa = 9.33IAVWRR118 pKa = 11.84DD119 pKa = 3.09YY120 pKa = 11.03FSWRR124 pKa = 11.84NVLLGAVTGTLRR136 pKa = 11.84SVRR139 pKa = 11.84GRR141 pKa = 11.84QRR143 pKa = 11.84HH144 pKa = 5.33LPAAPGHH151 pKa = 5.7GRR153 pKa = 11.84STGTIPGAAPATVPPDD169 pKa = 3.71YY170 pKa = 10.3PRR172 pKa = 11.84KK173 pKa = 9.88DD174 pKa = 3.34QVKK177 pKa = 9.87
Molecular weight: 19.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1634117 |
29 |
9173 |
327.7 |
34.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.428 ± 0.047 | 0.747 ± 0.011 |
6.2 ± 0.03 | 5.432 ± 0.035 |
2.964 ± 0.021 | 9.336 ± 0.037 |
2.055 ± 0.018 | 3.686 ± 0.026 |
1.983 ± 0.029 | 10.098 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.941 ± 0.015 | 2.005 ± 0.02 |
5.773 ± 0.031 | 2.736 ± 0.016 |
7.272 ± 0.04 | 5.466 ± 0.025 |
6.214 ± 0.032 | 9.215 ± 0.041 |
1.46 ± 0.014 | 1.99 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
