
Imjin virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Hantaviridae; Mammantavirinae; Thottimvirus; Imjin thottimvirus
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
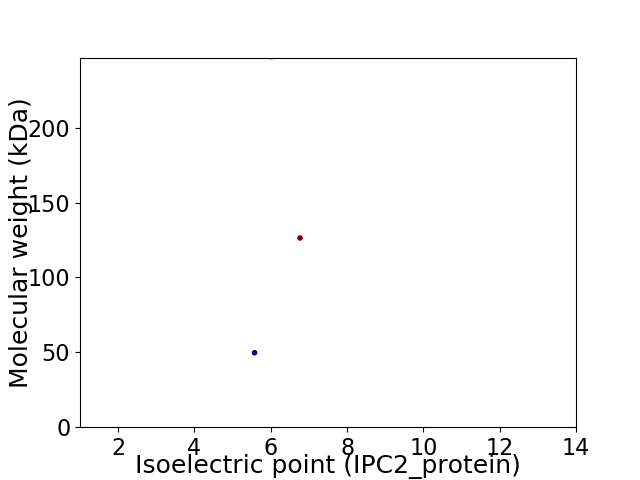
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
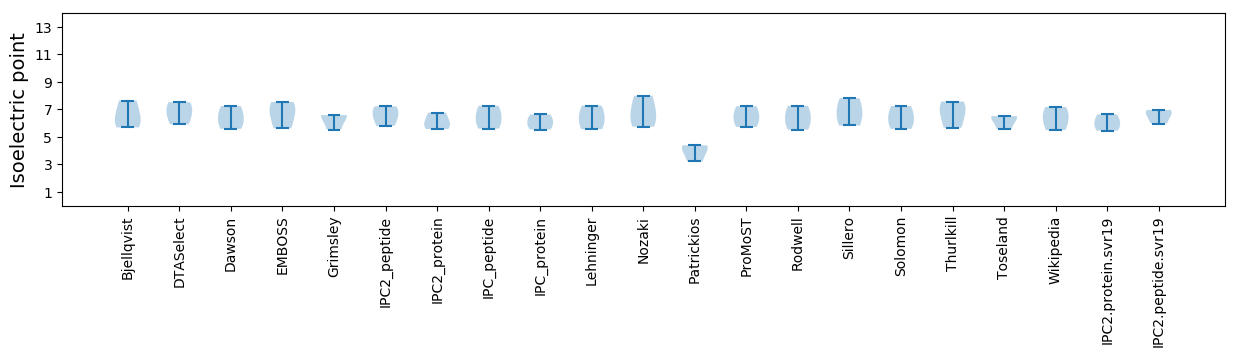
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075IK44|A0A075IK44_9VIRU Replicase OS=Imjin virus OX=467989 PE=4 SV=1
MM1 pKa = 6.87AQSRR5 pKa = 11.84ITRR8 pKa = 11.84EE9 pKa = 4.31DD10 pKa = 3.12IQKK13 pKa = 10.87LEE15 pKa = 4.13DD16 pKa = 3.25AKK18 pKa = 11.07KK19 pKa = 10.39QIEE22 pKa = 4.27IQLSSSIQKK31 pKa = 10.2LATEE35 pKa = 4.91TEE37 pKa = 4.38QFGPDD42 pKa = 3.8PDD44 pKa = 5.6PIQLQAVRR52 pKa = 11.84QRR54 pKa = 11.84SSVIEE59 pKa = 4.4GYY61 pKa = 9.73KK62 pKa = 10.44EE63 pKa = 3.74KK64 pKa = 10.92LRR66 pKa = 11.84DD67 pKa = 3.68LEE69 pKa = 4.43GQLSIAYY76 pKa = 7.64ATFADD81 pKa = 4.23EE82 pKa = 4.21EE83 pKa = 4.44EE84 pKa = 4.13EE85 pKa = 4.04RR86 pKa = 11.84KK87 pKa = 9.87KK88 pKa = 10.73KK89 pKa = 10.7RR90 pKa = 11.84EE91 pKa = 3.84VPGVEE96 pKa = 3.78EE97 pKa = 5.05GDD99 pKa = 3.83YY100 pKa = 11.11LSQKK104 pKa = 10.51SSLRR108 pKa = 11.84YY109 pKa = 10.4GNVIDD114 pKa = 5.82LNPLNLEE121 pKa = 4.1EE122 pKa = 5.2PAGQTANWNRR132 pKa = 11.84IFEE135 pKa = 4.6YY136 pKa = 10.11IATITQVLLLKK147 pKa = 10.48GLYY150 pKa = 9.91ILTTRR155 pKa = 11.84GRR157 pKa = 11.84QTSKK161 pKa = 10.99DD162 pKa = 3.61NKK164 pKa = 7.3GTRR167 pKa = 11.84MKK169 pKa = 10.89LKK171 pKa = 10.31DD172 pKa = 4.19DD173 pKa = 3.86SCMEE177 pKa = 4.57TINGIQRR184 pKa = 11.84HH185 pKa = 4.93KK186 pKa = 10.49FLYY189 pKa = 10.03ISLPTSQSSIQDD201 pKa = 3.63DD202 pKa = 4.15EE203 pKa = 4.38LTPGRR208 pKa = 11.84FRR210 pKa = 11.84TIVSGLLPNEE220 pKa = 4.18IKK222 pKa = 10.51SKK224 pKa = 11.03KK225 pKa = 10.11LMSPVMGVIGFLNLTEE241 pKa = 3.85VWMDD245 pKa = 3.68IIDD248 pKa = 5.36RR249 pKa = 11.84IMTDD253 pKa = 2.47QCDD256 pKa = 3.55YY257 pKa = 8.19MTQDD261 pKa = 3.57KK262 pKa = 11.07SNPTASTNRR271 pKa = 11.84AYY273 pKa = 10.54FKK275 pKa = 10.58EE276 pKa = 4.04RR277 pKa = 11.84QNTINSLNLPDD288 pKa = 5.17LKK290 pKa = 10.58QLRR293 pKa = 11.84TDD295 pKa = 3.93AEE297 pKa = 4.33RR298 pKa = 11.84KK299 pKa = 9.72HH300 pKa = 5.93YY301 pKa = 10.83LPDD304 pKa = 5.31DD305 pKa = 3.91IEE307 pKa = 4.63TEE309 pKa = 4.21CVPWIFANAPDD320 pKa = 4.11RR321 pKa = 11.84CPPTVLLVAGIPEE334 pKa = 4.66LGAFFALMQDD344 pKa = 2.92IRR346 pKa = 11.84SGILASNLRR355 pKa = 11.84GTAEE359 pKa = 3.79EE360 pKa = 4.37KK361 pKa = 10.25IARR364 pKa = 11.84KK365 pKa = 9.81SSFYY369 pKa = 10.31QSYY372 pKa = 10.27IRR374 pKa = 11.84RR375 pKa = 11.84TQSMGLNCDD384 pKa = 2.99QKK386 pKa = 10.76IIHH389 pKa = 7.06IYY391 pKa = 9.25MDD393 pKa = 3.71YY394 pKa = 11.21LGTFCVDD401 pKa = 3.59HH402 pKa = 6.9FNLGDD407 pKa = 5.38DD408 pKa = 4.26MDD410 pKa = 5.66PDD412 pKa = 4.46LKK414 pKa = 10.77IKK416 pKa = 10.53AQALLDD422 pKa = 3.93KK423 pKa = 10.22KK424 pKa = 10.79VKK426 pKa = 10.2EE427 pKa = 4.29ISTQEE432 pKa = 4.13PIKK435 pKa = 10.86LL436 pKa = 3.59
MM1 pKa = 6.87AQSRR5 pKa = 11.84ITRR8 pKa = 11.84EE9 pKa = 4.31DD10 pKa = 3.12IQKK13 pKa = 10.87LEE15 pKa = 4.13DD16 pKa = 3.25AKK18 pKa = 11.07KK19 pKa = 10.39QIEE22 pKa = 4.27IQLSSSIQKK31 pKa = 10.2LATEE35 pKa = 4.91TEE37 pKa = 4.38QFGPDD42 pKa = 3.8PDD44 pKa = 5.6PIQLQAVRR52 pKa = 11.84QRR54 pKa = 11.84SSVIEE59 pKa = 4.4GYY61 pKa = 9.73KK62 pKa = 10.44EE63 pKa = 3.74KK64 pKa = 10.92LRR66 pKa = 11.84DD67 pKa = 3.68LEE69 pKa = 4.43GQLSIAYY76 pKa = 7.64ATFADD81 pKa = 4.23EE82 pKa = 4.21EE83 pKa = 4.44EE84 pKa = 4.13EE85 pKa = 4.04RR86 pKa = 11.84KK87 pKa = 9.87KK88 pKa = 10.73KK89 pKa = 10.7RR90 pKa = 11.84EE91 pKa = 3.84VPGVEE96 pKa = 3.78EE97 pKa = 5.05GDD99 pKa = 3.83YY100 pKa = 11.11LSQKK104 pKa = 10.51SSLRR108 pKa = 11.84YY109 pKa = 10.4GNVIDD114 pKa = 5.82LNPLNLEE121 pKa = 4.1EE122 pKa = 5.2PAGQTANWNRR132 pKa = 11.84IFEE135 pKa = 4.6YY136 pKa = 10.11IATITQVLLLKK147 pKa = 10.48GLYY150 pKa = 9.91ILTTRR155 pKa = 11.84GRR157 pKa = 11.84QTSKK161 pKa = 10.99DD162 pKa = 3.61NKK164 pKa = 7.3GTRR167 pKa = 11.84MKK169 pKa = 10.89LKK171 pKa = 10.31DD172 pKa = 4.19DD173 pKa = 3.86SCMEE177 pKa = 4.57TINGIQRR184 pKa = 11.84HH185 pKa = 4.93KK186 pKa = 10.49FLYY189 pKa = 10.03ISLPTSQSSIQDD201 pKa = 3.63DD202 pKa = 4.15EE203 pKa = 4.38LTPGRR208 pKa = 11.84FRR210 pKa = 11.84TIVSGLLPNEE220 pKa = 4.18IKK222 pKa = 10.51SKK224 pKa = 11.03KK225 pKa = 10.11LMSPVMGVIGFLNLTEE241 pKa = 3.85VWMDD245 pKa = 3.68IIDD248 pKa = 5.36RR249 pKa = 11.84IMTDD253 pKa = 2.47QCDD256 pKa = 3.55YY257 pKa = 8.19MTQDD261 pKa = 3.57KK262 pKa = 11.07SNPTASTNRR271 pKa = 11.84AYY273 pKa = 10.54FKK275 pKa = 10.58EE276 pKa = 4.04RR277 pKa = 11.84QNTINSLNLPDD288 pKa = 5.17LKK290 pKa = 10.58QLRR293 pKa = 11.84TDD295 pKa = 3.93AEE297 pKa = 4.33RR298 pKa = 11.84KK299 pKa = 9.72HH300 pKa = 5.93YY301 pKa = 10.83LPDD304 pKa = 5.31DD305 pKa = 3.91IEE307 pKa = 4.63TEE309 pKa = 4.21CVPWIFANAPDD320 pKa = 4.11RR321 pKa = 11.84CPPTVLLVAGIPEE334 pKa = 4.66LGAFFALMQDD344 pKa = 2.92IRR346 pKa = 11.84SGILASNLRR355 pKa = 11.84GTAEE359 pKa = 3.79EE360 pKa = 4.37KK361 pKa = 10.25IARR364 pKa = 11.84KK365 pKa = 9.81SSFYY369 pKa = 10.31QSYY372 pKa = 10.27IRR374 pKa = 11.84RR375 pKa = 11.84TQSMGLNCDD384 pKa = 2.99QKK386 pKa = 10.76IIHH389 pKa = 7.06IYY391 pKa = 9.25MDD393 pKa = 3.71YY394 pKa = 11.21LGTFCVDD401 pKa = 3.59HH402 pKa = 6.9FNLGDD407 pKa = 5.38DD408 pKa = 4.26MDD410 pKa = 5.66PDD412 pKa = 4.46LKK414 pKa = 10.77IKK416 pKa = 10.53AQALLDD422 pKa = 3.93KK423 pKa = 10.22KK424 pKa = 10.79VKK426 pKa = 10.2EE427 pKa = 4.29ISTQEE432 pKa = 4.13PIKK435 pKa = 10.86LL436 pKa = 3.59
Molecular weight: 49.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075II01|A0A075II01_9VIRU Nucleocapsid protein OS=Imjin virus OX=467989 PE=3 SV=1
MM1 pKa = 7.46NNGFIIVLLQSLLQVYY17 pKa = 10.0CSTHH21 pKa = 5.54SRR23 pKa = 11.84ITLEE27 pKa = 4.17CPHH30 pKa = 7.08YY31 pKa = 10.28GVSDD35 pKa = 3.85AEE37 pKa = 4.46YY38 pKa = 9.21VWGKK42 pKa = 10.16VKK44 pKa = 10.43IGRR47 pKa = 11.84FTIDD51 pKa = 4.48DD52 pKa = 3.44IKK54 pKa = 10.93HH55 pKa = 5.21IQFEE59 pKa = 4.59NTCPFSITGHH69 pKa = 5.32NVQTGMKK76 pKa = 9.15IISWTEE82 pKa = 3.8KK83 pKa = 10.19EE84 pKa = 4.21NHH86 pKa = 6.97DD87 pKa = 3.8SSAVYY92 pKa = 9.41FDD94 pKa = 4.33KK95 pKa = 11.28VATHH99 pKa = 5.87SEE101 pKa = 3.88KK102 pKa = 10.49TGDD105 pKa = 3.76FKK107 pKa = 11.43SLCGSVSYY115 pKa = 8.86LTYY118 pKa = 9.87PFKK121 pKa = 11.05SGSSYY126 pKa = 11.08CLNLHH131 pKa = 6.86CNQTVCDD138 pKa = 3.79TKK140 pKa = 11.22VIVLIKK146 pKa = 10.19AISWVAYY153 pKa = 9.0DD154 pKa = 4.13SCVLYY159 pKa = 10.92HH160 pKa = 7.21KK161 pKa = 10.11DD162 pKa = 3.59IKK164 pKa = 10.89VQLDD168 pKa = 3.74FKK170 pKa = 10.23KK171 pKa = 7.54THH173 pKa = 5.8CVKK176 pKa = 10.73SVLHH180 pKa = 6.09EE181 pKa = 4.4KK182 pKa = 10.5KK183 pKa = 10.45CFQPDD188 pKa = 3.14FTTAFNPKK196 pKa = 9.36LHH198 pKa = 6.59MKK200 pKa = 10.39FSLPSVCLFMPSDD213 pKa = 3.9GNAEE217 pKa = 4.02GTKK220 pKa = 10.09EE221 pKa = 3.96IVGQDD226 pKa = 2.96SCTDD230 pKa = 2.86IKK232 pKa = 11.36LNGWYY237 pKa = 9.93SCIVGHH243 pKa = 6.02YY244 pKa = 11.07SMIFVHH250 pKa = 6.73SNGKK254 pKa = 8.06MRR256 pKa = 11.84QIHH259 pKa = 6.8LNTKK263 pKa = 8.53FRR265 pKa = 11.84KK266 pKa = 9.29NPQGDD271 pKa = 3.59DD272 pKa = 3.44HH273 pKa = 8.55SEE275 pKa = 3.87LRR277 pKa = 11.84SGSTHH282 pKa = 6.62IIGYY286 pKa = 10.49KK287 pKa = 10.68NMTTVSNKK295 pKa = 9.66VNTLAAFKK303 pKa = 10.73GATDD307 pKa = 3.9YY308 pKa = 11.8SSLFYY313 pKa = 10.85NIQNTDD319 pKa = 3.25KK320 pKa = 11.15KK321 pKa = 10.04PLLLAGYY328 pKa = 7.5TPTFHH333 pKa = 6.82WRR335 pKa = 11.84EE336 pKa = 4.38CGTHH340 pKa = 7.38LIPAVWTSHH349 pKa = 5.35IDD351 pKa = 3.1VWGTIIEE358 pKa = 4.13SDD360 pKa = 3.64EE361 pKa = 4.37CNIFCTLSGPGASCEE376 pKa = 4.11AFSPTGIFLLNSTTCLIPHH395 pKa = 5.49THH397 pKa = 6.41RR398 pKa = 11.84FKK400 pKa = 11.42GLGDD404 pKa = 3.85QITFICQRR412 pKa = 11.84VDD414 pKa = 2.86TDD416 pKa = 5.05LEE418 pKa = 5.04FYY420 pKa = 11.17CNGRR424 pKa = 11.84HH425 pKa = 6.35KK426 pKa = 10.52IIRR429 pKa = 11.84TKK431 pKa = 9.94TLIIGQCIQTFTSFFSLFPGVAHH454 pKa = 6.56SLAVEE459 pKa = 4.11LCVPGFHH466 pKa = 7.18GFITVCIVSTFCFGWLWIPGLTWLILQLLKK496 pKa = 10.89FIVLFVSTNSVDD508 pKa = 3.57QRR510 pKa = 11.84FKK512 pKa = 11.25QILQKK517 pKa = 10.56IKK519 pKa = 10.54AEE521 pKa = 3.92YY522 pKa = 10.05RR523 pKa = 11.84LTIGDD528 pKa = 4.32TSCEE532 pKa = 4.09FCKK535 pKa = 10.71HH536 pKa = 5.05EE537 pKa = 4.59CQTSLEE543 pKa = 4.26SDD545 pKa = 3.51SHH547 pKa = 8.4ILYY550 pKa = 10.09CKK552 pKa = 9.88QGKK555 pKa = 9.1CPYY558 pKa = 8.94CLNEE562 pKa = 4.61IYY564 pKa = 10.2PSPLALQEE572 pKa = 4.18HH573 pKa = 6.68YY574 pKa = 9.62KK575 pKa = 10.61VCMLTDD581 pKa = 3.66RR582 pKa = 11.84FTRR585 pKa = 11.84QIKK588 pKa = 8.82EE589 pKa = 3.74KK590 pKa = 10.47IMNVEE595 pKa = 4.51TKK597 pKa = 9.81PHH599 pKa = 5.54SGLYY603 pKa = 9.94KK604 pKa = 10.09RR605 pKa = 11.84LCVFQYY611 pKa = 11.15KK612 pKa = 9.24NRR614 pKa = 11.84CYY616 pKa = 10.55IFTIWIFLLIFQMLVWAASAEE637 pKa = 4.29VITFEE642 pKa = 5.64KK643 pKa = 10.04EE644 pKa = 3.69WNDD647 pKa = 3.24NVHH650 pKa = 6.96GIGSEE655 pKa = 3.95KK656 pKa = 10.75LITDD660 pKa = 4.42LEE662 pKa = 4.66LDD664 pKa = 3.99FSIPSSSGFTYY675 pKa = 9.92TRR677 pKa = 11.84NLEE680 pKa = 4.12GPIPNQKK687 pKa = 9.83LHH689 pKa = 6.44FALKK693 pKa = 10.66LSRR696 pKa = 11.84QKK698 pKa = 9.5TRR700 pKa = 11.84ATIQKK705 pKa = 9.39LGHH708 pKa = 6.01WVDD711 pKa = 3.52ARR713 pKa = 11.84WNVRR717 pKa = 11.84TVFHH721 pKa = 7.23CYY723 pKa = 9.22GACSKK728 pKa = 10.94FEE730 pKa = 4.22YY731 pKa = 8.91PWKK734 pKa = 10.55SATCSRR740 pKa = 11.84EE741 pKa = 3.36KK742 pKa = 10.88DD743 pKa = 3.41FEE745 pKa = 4.35YY746 pKa = 9.64QTGWGCNPGDD756 pKa = 3.99CPGINTGCTACGLYY770 pKa = 10.18LDD772 pKa = 4.82KK773 pKa = 10.53PRR775 pKa = 11.84SIATVIKK782 pKa = 10.34LIQLDD787 pKa = 4.04YY788 pKa = 10.87EE789 pKa = 4.41RR790 pKa = 11.84EE791 pKa = 3.96VCIQIGTHH799 pKa = 5.46SEE801 pKa = 4.16CKK803 pKa = 10.37KK804 pKa = 9.65ITGNDD809 pKa = 3.49CLSTHH814 pKa = 5.79GVKK817 pKa = 9.68ICLLSTTVKK826 pKa = 9.67LTATDD831 pKa = 3.47TLVFFGPLQQGAVVFKK847 pKa = 10.05NWCTSTCAYY856 pKa = 9.95GDD858 pKa = 4.14PGDD861 pKa = 5.04IMLTEE866 pKa = 4.28NGEE869 pKa = 4.35YY870 pKa = 10.31NCPDD874 pKa = 3.1FTGSFEE880 pKa = 4.12RR881 pKa = 11.84TCRR884 pKa = 11.84FGQTPVCEE892 pKa = 4.09YY893 pKa = 10.44NGNNIGGYY901 pKa = 9.34KK902 pKa = 9.97RR903 pKa = 11.84YY904 pKa = 9.62LATRR908 pKa = 11.84DD909 pKa = 3.44SFFSINMTEE918 pKa = 4.17PIIDD922 pKa = 3.63KK923 pKa = 10.3TKK925 pKa = 11.13LEE927 pKa = 4.14WFDD930 pKa = 4.11PDD932 pKa = 3.55STSRR936 pKa = 11.84DD937 pKa = 3.64HH938 pKa = 8.15INVQVSKK945 pKa = 11.21DD946 pKa = 3.29VDD948 pKa = 4.19FEE950 pKa = 4.44NLGNNPCRR958 pKa = 11.84LTLKK962 pKa = 7.86TTGIEE967 pKa = 4.26GAWGSEE973 pKa = 3.84TGFSLTCKK981 pKa = 10.34ISLLEE986 pKa = 4.3CNSFVTMIKK995 pKa = 10.38ACDD998 pKa = 3.18KK999 pKa = 10.71AMCYY1003 pKa = 9.84GAYY1006 pKa = 8.76STTLQRR1012 pKa = 11.84GDD1014 pKa = 3.65NNVIIQGRR1022 pKa = 11.84GGHH1025 pKa = 6.0SGSSFKK1031 pKa = 10.81CCHH1034 pKa = 6.52NDD1036 pKa = 3.25DD1037 pKa = 4.21CSEE1040 pKa = 4.13TGVLADD1046 pKa = 4.31APHH1049 pKa = 6.97LSRR1052 pKa = 11.84LKK1054 pKa = 10.98SQDD1057 pKa = 2.83GDD1059 pKa = 3.36NSEE1062 pKa = 4.62IYY1064 pKa = 10.57SDD1066 pKa = 3.84GANEE1070 pKa = 4.57CRR1072 pKa = 11.84ITCWFNKK1079 pKa = 7.63TSEE1082 pKa = 4.12WVMGMLSGNWLVILVLVGIMLFSVFLFCFFCPAKK1116 pKa = 9.62THH1118 pKa = 5.42QAA1120 pKa = 3.31
MM1 pKa = 7.46NNGFIIVLLQSLLQVYY17 pKa = 10.0CSTHH21 pKa = 5.54SRR23 pKa = 11.84ITLEE27 pKa = 4.17CPHH30 pKa = 7.08YY31 pKa = 10.28GVSDD35 pKa = 3.85AEE37 pKa = 4.46YY38 pKa = 9.21VWGKK42 pKa = 10.16VKK44 pKa = 10.43IGRR47 pKa = 11.84FTIDD51 pKa = 4.48DD52 pKa = 3.44IKK54 pKa = 10.93HH55 pKa = 5.21IQFEE59 pKa = 4.59NTCPFSITGHH69 pKa = 5.32NVQTGMKK76 pKa = 9.15IISWTEE82 pKa = 3.8KK83 pKa = 10.19EE84 pKa = 4.21NHH86 pKa = 6.97DD87 pKa = 3.8SSAVYY92 pKa = 9.41FDD94 pKa = 4.33KK95 pKa = 11.28VATHH99 pKa = 5.87SEE101 pKa = 3.88KK102 pKa = 10.49TGDD105 pKa = 3.76FKK107 pKa = 11.43SLCGSVSYY115 pKa = 8.86LTYY118 pKa = 9.87PFKK121 pKa = 11.05SGSSYY126 pKa = 11.08CLNLHH131 pKa = 6.86CNQTVCDD138 pKa = 3.79TKK140 pKa = 11.22VIVLIKK146 pKa = 10.19AISWVAYY153 pKa = 9.0DD154 pKa = 4.13SCVLYY159 pKa = 10.92HH160 pKa = 7.21KK161 pKa = 10.11DD162 pKa = 3.59IKK164 pKa = 10.89VQLDD168 pKa = 3.74FKK170 pKa = 10.23KK171 pKa = 7.54THH173 pKa = 5.8CVKK176 pKa = 10.73SVLHH180 pKa = 6.09EE181 pKa = 4.4KK182 pKa = 10.5KK183 pKa = 10.45CFQPDD188 pKa = 3.14FTTAFNPKK196 pKa = 9.36LHH198 pKa = 6.59MKK200 pKa = 10.39FSLPSVCLFMPSDD213 pKa = 3.9GNAEE217 pKa = 4.02GTKK220 pKa = 10.09EE221 pKa = 3.96IVGQDD226 pKa = 2.96SCTDD230 pKa = 2.86IKK232 pKa = 11.36LNGWYY237 pKa = 9.93SCIVGHH243 pKa = 6.02YY244 pKa = 11.07SMIFVHH250 pKa = 6.73SNGKK254 pKa = 8.06MRR256 pKa = 11.84QIHH259 pKa = 6.8LNTKK263 pKa = 8.53FRR265 pKa = 11.84KK266 pKa = 9.29NPQGDD271 pKa = 3.59DD272 pKa = 3.44HH273 pKa = 8.55SEE275 pKa = 3.87LRR277 pKa = 11.84SGSTHH282 pKa = 6.62IIGYY286 pKa = 10.49KK287 pKa = 10.68NMTTVSNKK295 pKa = 9.66VNTLAAFKK303 pKa = 10.73GATDD307 pKa = 3.9YY308 pKa = 11.8SSLFYY313 pKa = 10.85NIQNTDD319 pKa = 3.25KK320 pKa = 11.15KK321 pKa = 10.04PLLLAGYY328 pKa = 7.5TPTFHH333 pKa = 6.82WRR335 pKa = 11.84EE336 pKa = 4.38CGTHH340 pKa = 7.38LIPAVWTSHH349 pKa = 5.35IDD351 pKa = 3.1VWGTIIEE358 pKa = 4.13SDD360 pKa = 3.64EE361 pKa = 4.37CNIFCTLSGPGASCEE376 pKa = 4.11AFSPTGIFLLNSTTCLIPHH395 pKa = 5.49THH397 pKa = 6.41RR398 pKa = 11.84FKK400 pKa = 11.42GLGDD404 pKa = 3.85QITFICQRR412 pKa = 11.84VDD414 pKa = 2.86TDD416 pKa = 5.05LEE418 pKa = 5.04FYY420 pKa = 11.17CNGRR424 pKa = 11.84HH425 pKa = 6.35KK426 pKa = 10.52IIRR429 pKa = 11.84TKK431 pKa = 9.94TLIIGQCIQTFTSFFSLFPGVAHH454 pKa = 6.56SLAVEE459 pKa = 4.11LCVPGFHH466 pKa = 7.18GFITVCIVSTFCFGWLWIPGLTWLILQLLKK496 pKa = 10.89FIVLFVSTNSVDD508 pKa = 3.57QRR510 pKa = 11.84FKK512 pKa = 11.25QILQKK517 pKa = 10.56IKK519 pKa = 10.54AEE521 pKa = 3.92YY522 pKa = 10.05RR523 pKa = 11.84LTIGDD528 pKa = 4.32TSCEE532 pKa = 4.09FCKK535 pKa = 10.71HH536 pKa = 5.05EE537 pKa = 4.59CQTSLEE543 pKa = 4.26SDD545 pKa = 3.51SHH547 pKa = 8.4ILYY550 pKa = 10.09CKK552 pKa = 9.88QGKK555 pKa = 9.1CPYY558 pKa = 8.94CLNEE562 pKa = 4.61IYY564 pKa = 10.2PSPLALQEE572 pKa = 4.18HH573 pKa = 6.68YY574 pKa = 9.62KK575 pKa = 10.61VCMLTDD581 pKa = 3.66RR582 pKa = 11.84FTRR585 pKa = 11.84QIKK588 pKa = 8.82EE589 pKa = 3.74KK590 pKa = 10.47IMNVEE595 pKa = 4.51TKK597 pKa = 9.81PHH599 pKa = 5.54SGLYY603 pKa = 9.94KK604 pKa = 10.09RR605 pKa = 11.84LCVFQYY611 pKa = 11.15KK612 pKa = 9.24NRR614 pKa = 11.84CYY616 pKa = 10.55IFTIWIFLLIFQMLVWAASAEE637 pKa = 4.29VITFEE642 pKa = 5.64KK643 pKa = 10.04EE644 pKa = 3.69WNDD647 pKa = 3.24NVHH650 pKa = 6.96GIGSEE655 pKa = 3.95KK656 pKa = 10.75LITDD660 pKa = 4.42LEE662 pKa = 4.66LDD664 pKa = 3.99FSIPSSSGFTYY675 pKa = 9.92TRR677 pKa = 11.84NLEE680 pKa = 4.12GPIPNQKK687 pKa = 9.83LHH689 pKa = 6.44FALKK693 pKa = 10.66LSRR696 pKa = 11.84QKK698 pKa = 9.5TRR700 pKa = 11.84ATIQKK705 pKa = 9.39LGHH708 pKa = 6.01WVDD711 pKa = 3.52ARR713 pKa = 11.84WNVRR717 pKa = 11.84TVFHH721 pKa = 7.23CYY723 pKa = 9.22GACSKK728 pKa = 10.94FEE730 pKa = 4.22YY731 pKa = 8.91PWKK734 pKa = 10.55SATCSRR740 pKa = 11.84EE741 pKa = 3.36KK742 pKa = 10.88DD743 pKa = 3.41FEE745 pKa = 4.35YY746 pKa = 9.64QTGWGCNPGDD756 pKa = 3.99CPGINTGCTACGLYY770 pKa = 10.18LDD772 pKa = 4.82KK773 pKa = 10.53PRR775 pKa = 11.84SIATVIKK782 pKa = 10.34LIQLDD787 pKa = 4.04YY788 pKa = 10.87EE789 pKa = 4.41RR790 pKa = 11.84EE791 pKa = 3.96VCIQIGTHH799 pKa = 5.46SEE801 pKa = 4.16CKK803 pKa = 10.37KK804 pKa = 9.65ITGNDD809 pKa = 3.49CLSTHH814 pKa = 5.79GVKK817 pKa = 9.68ICLLSTTVKK826 pKa = 9.67LTATDD831 pKa = 3.47TLVFFGPLQQGAVVFKK847 pKa = 10.05NWCTSTCAYY856 pKa = 9.95GDD858 pKa = 4.14PGDD861 pKa = 5.04IMLTEE866 pKa = 4.28NGEE869 pKa = 4.35YY870 pKa = 10.31NCPDD874 pKa = 3.1FTGSFEE880 pKa = 4.12RR881 pKa = 11.84TCRR884 pKa = 11.84FGQTPVCEE892 pKa = 4.09YY893 pKa = 10.44NGNNIGGYY901 pKa = 9.34KK902 pKa = 9.97RR903 pKa = 11.84YY904 pKa = 9.62LATRR908 pKa = 11.84DD909 pKa = 3.44SFFSINMTEE918 pKa = 4.17PIIDD922 pKa = 3.63KK923 pKa = 10.3TKK925 pKa = 11.13LEE927 pKa = 4.14WFDD930 pKa = 4.11PDD932 pKa = 3.55STSRR936 pKa = 11.84DD937 pKa = 3.64HH938 pKa = 8.15INVQVSKK945 pKa = 11.21DD946 pKa = 3.29VDD948 pKa = 4.19FEE950 pKa = 4.44NLGNNPCRR958 pKa = 11.84LTLKK962 pKa = 7.86TTGIEE967 pKa = 4.26GAWGSEE973 pKa = 3.84TGFSLTCKK981 pKa = 10.34ISLLEE986 pKa = 4.3CNSFVTMIKK995 pKa = 10.38ACDD998 pKa = 3.18KK999 pKa = 10.71AMCYY1003 pKa = 9.84GAYY1006 pKa = 8.76STTLQRR1012 pKa = 11.84GDD1014 pKa = 3.65NNVIIQGRR1022 pKa = 11.84GGHH1025 pKa = 6.0SGSSFKK1031 pKa = 10.81CCHH1034 pKa = 6.52NDD1036 pKa = 3.25DD1037 pKa = 4.21CSEE1040 pKa = 4.13TGVLADD1046 pKa = 4.31APHH1049 pKa = 6.97LSRR1052 pKa = 11.84LKK1054 pKa = 10.98SQDD1057 pKa = 2.83GDD1059 pKa = 3.36NSEE1062 pKa = 4.62IYY1064 pKa = 10.57SDD1066 pKa = 3.84GANEE1070 pKa = 4.57CRR1072 pKa = 11.84ITCWFNKK1079 pKa = 7.63TSEE1082 pKa = 4.12WVMGMLSGNWLVILVLVGIMLFSVFLFCFFCPAKK1116 pKa = 9.62THH1118 pKa = 5.42QAA1120 pKa = 3.31
Molecular weight: 126.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3705 |
436 |
2149 |
1235.0 |
140.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.723 ± 0.456 | 2.699 ± 1.179 |
5.668 ± 0.515 | 6.019 ± 0.676 |
5.155 ± 0.648 | 5.479 ± 0.682 |
2.618 ± 0.541 | 7.368 ± 0.325 |
7.152 ± 0.235 | 9.474 ± 0.54 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.97 ± 0.26 | 4.534 ± 0.117 |
3.59 ± 0.29 | 4.238 ± 0.584 |
4.022 ± 0.617 | 7.26 ± 0.219 |
6.478 ± 0.82 | 6.046 ± 0.891 |
1.592 ± 0.288 | 3.914 ± 0.364 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
