
Desulfobacterium vacuolatum DSM 3385
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Desulfobacterium; Desulfobacterium vacuolatum
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
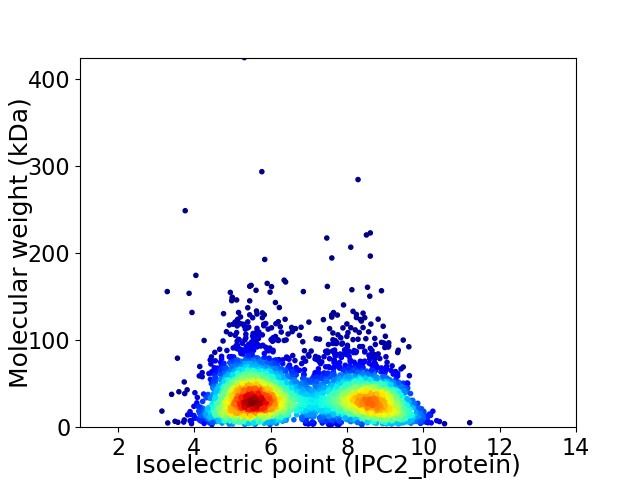
Virtual 2D-PAGE plot for 4083 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W1YN33|A0A1W1YN33_9DELT Acetoin utilization protein AcuB OS=Desulfobacterium vacuolatum DSM 3385 OX=1121400 GN=SAMN02746065_101204 PE=4 SV=1
MM1 pKa = 7.56KK2 pKa = 10.68KK3 pKa = 8.27LTRR6 pKa = 11.84YY7 pKa = 9.09IAIAVTCLFFVAAGANAMTFKK28 pKa = 10.74AGDD31 pKa = 3.59MGEE34 pKa = 4.15MQINAAFKK42 pKa = 10.53ASFGATQYY50 pKa = 11.72LDD52 pKa = 4.35FVDD55 pKa = 5.85DD56 pKa = 3.92YY57 pKa = 12.07SKK59 pKa = 11.79GDD61 pKa = 4.02DD62 pKa = 3.67SDD64 pKa = 4.89FDD66 pKa = 3.74ATRR69 pKa = 11.84EE70 pKa = 4.01LSFKK74 pKa = 10.61AQWILNKK81 pKa = 9.82NLKK84 pKa = 10.3GVVSFQIGEE93 pKa = 4.31GSTGGYY99 pKa = 9.76FGSTDD104 pKa = 3.21ALVGGEE110 pKa = 4.14EE111 pKa = 4.42DD112 pKa = 3.7GDD114 pKa = 5.05LILEE118 pKa = 4.74LDD120 pKa = 3.54NLYY123 pKa = 10.18IDD125 pKa = 4.52FTTDD129 pKa = 2.35SGINFKK135 pKa = 10.42IGSQGFSLAEE145 pKa = 3.47IAYY148 pKa = 9.36GSNLFYY154 pKa = 10.47EE155 pKa = 5.03VPAGVTFSAPLTNSSSLQAGWFRR178 pKa = 11.84MADD181 pKa = 3.51LMDD184 pKa = 5.27DD185 pKa = 4.17SNSNTDD191 pKa = 3.34DD192 pKa = 3.54QADD195 pKa = 3.64FLYY198 pKa = 10.96AKK200 pKa = 10.4LPVKK204 pKa = 9.96MDD206 pKa = 5.3AISFTPWAAYY216 pKa = 10.52ANIQEE221 pKa = 4.63DD222 pKa = 4.68VIANAPSHH230 pKa = 5.25WRR232 pKa = 11.84YY233 pKa = 10.45AYY235 pKa = 10.2FDD237 pKa = 3.72YY238 pKa = 11.11PGFLTGANSAVIDD251 pKa = 3.84PTATDD256 pKa = 5.46DD257 pKa = 3.64VSAWYY262 pKa = 10.24LGVSFGYY269 pKa = 9.62TMDD272 pKa = 4.21ALSIQASATYY282 pKa = 11.08GDD284 pKa = 4.69MDD286 pKa = 4.03WEE288 pKa = 4.42TATVDD293 pKa = 3.62ASIAGYY299 pKa = 9.29FADD302 pKa = 5.56LVIDD306 pKa = 3.69YY307 pKa = 10.67KK308 pKa = 10.64MDD310 pKa = 3.21GFTPEE315 pKa = 4.27FFAFYY320 pKa = 11.02GNGPDD325 pKa = 5.52ANDD328 pKa = 3.74EE329 pKa = 4.61DD330 pKa = 5.9FDD332 pKa = 4.13MMPVLIGGPTYY343 pKa = 9.7TSSYY347 pKa = 10.28FGGSRR352 pKa = 11.84FNDD355 pKa = 3.34NMFDD359 pKa = 4.34SYY361 pKa = 11.8DD362 pKa = 3.52STYY365 pKa = 9.48ATSMWAVGFKK375 pKa = 10.86VKK377 pKa = 10.4DD378 pKa = 3.19IKK380 pKa = 10.27TGEE383 pKa = 4.22KK384 pKa = 10.05LSHH387 pKa = 6.67EE388 pKa = 4.33FQIMYY393 pKa = 10.77AEE395 pKa = 4.62GTAEE399 pKa = 3.85DD400 pKa = 5.01TIFQSPNDD408 pKa = 3.36ILMNEE413 pKa = 4.39DD414 pKa = 3.38EE415 pKa = 5.54SVLEE419 pKa = 4.06VNFNSEE425 pKa = 4.12YY426 pKa = 10.83QIMKK430 pKa = 9.65HH431 pKa = 5.92LLFATEE437 pKa = 4.53LGYY440 pKa = 7.79MTFDD444 pKa = 3.59EE445 pKa = 5.08DD446 pKa = 3.94SDD448 pKa = 4.31YY449 pKa = 11.94NEE451 pKa = 4.51ALNGSVEE458 pKa = 4.58DD459 pKa = 4.18FWKK462 pKa = 10.24VACAIEE468 pKa = 4.5LSFF471 pKa = 5.5
MM1 pKa = 7.56KK2 pKa = 10.68KK3 pKa = 8.27LTRR6 pKa = 11.84YY7 pKa = 9.09IAIAVTCLFFVAAGANAMTFKK28 pKa = 10.74AGDD31 pKa = 3.59MGEE34 pKa = 4.15MQINAAFKK42 pKa = 10.53ASFGATQYY50 pKa = 11.72LDD52 pKa = 4.35FVDD55 pKa = 5.85DD56 pKa = 3.92YY57 pKa = 12.07SKK59 pKa = 11.79GDD61 pKa = 4.02DD62 pKa = 3.67SDD64 pKa = 4.89FDD66 pKa = 3.74ATRR69 pKa = 11.84EE70 pKa = 4.01LSFKK74 pKa = 10.61AQWILNKK81 pKa = 9.82NLKK84 pKa = 10.3GVVSFQIGEE93 pKa = 4.31GSTGGYY99 pKa = 9.76FGSTDD104 pKa = 3.21ALVGGEE110 pKa = 4.14EE111 pKa = 4.42DD112 pKa = 3.7GDD114 pKa = 5.05LILEE118 pKa = 4.74LDD120 pKa = 3.54NLYY123 pKa = 10.18IDD125 pKa = 4.52FTTDD129 pKa = 2.35SGINFKK135 pKa = 10.42IGSQGFSLAEE145 pKa = 3.47IAYY148 pKa = 9.36GSNLFYY154 pKa = 10.47EE155 pKa = 5.03VPAGVTFSAPLTNSSSLQAGWFRR178 pKa = 11.84MADD181 pKa = 3.51LMDD184 pKa = 5.27DD185 pKa = 4.17SNSNTDD191 pKa = 3.34DD192 pKa = 3.54QADD195 pKa = 3.64FLYY198 pKa = 10.96AKK200 pKa = 10.4LPVKK204 pKa = 9.96MDD206 pKa = 5.3AISFTPWAAYY216 pKa = 10.52ANIQEE221 pKa = 4.63DD222 pKa = 4.68VIANAPSHH230 pKa = 5.25WRR232 pKa = 11.84YY233 pKa = 10.45AYY235 pKa = 10.2FDD237 pKa = 3.72YY238 pKa = 11.11PGFLTGANSAVIDD251 pKa = 3.84PTATDD256 pKa = 5.46DD257 pKa = 3.64VSAWYY262 pKa = 10.24LGVSFGYY269 pKa = 9.62TMDD272 pKa = 4.21ALSIQASATYY282 pKa = 11.08GDD284 pKa = 4.69MDD286 pKa = 4.03WEE288 pKa = 4.42TATVDD293 pKa = 3.62ASIAGYY299 pKa = 9.29FADD302 pKa = 5.56LVIDD306 pKa = 3.69YY307 pKa = 10.67KK308 pKa = 10.64MDD310 pKa = 3.21GFTPEE315 pKa = 4.27FFAFYY320 pKa = 11.02GNGPDD325 pKa = 5.52ANDD328 pKa = 3.74EE329 pKa = 4.61DD330 pKa = 5.9FDD332 pKa = 4.13MMPVLIGGPTYY343 pKa = 9.7TSSYY347 pKa = 10.28FGGSRR352 pKa = 11.84FNDD355 pKa = 3.34NMFDD359 pKa = 4.34SYY361 pKa = 11.8DD362 pKa = 3.52STYY365 pKa = 9.48ATSMWAVGFKK375 pKa = 10.86VKK377 pKa = 10.4DD378 pKa = 3.19IKK380 pKa = 10.27TGEE383 pKa = 4.22KK384 pKa = 10.05LSHH387 pKa = 6.67EE388 pKa = 4.33FQIMYY393 pKa = 10.77AEE395 pKa = 4.62GTAEE399 pKa = 3.85DD400 pKa = 5.01TIFQSPNDD408 pKa = 3.36ILMNEE413 pKa = 4.39DD414 pKa = 3.38EE415 pKa = 5.54SVLEE419 pKa = 4.06VNFNSEE425 pKa = 4.12YY426 pKa = 10.83QIMKK430 pKa = 9.65HH431 pKa = 5.92LLFATEE437 pKa = 4.53LGYY440 pKa = 7.79MTFDD444 pKa = 3.59EE445 pKa = 5.08DD446 pKa = 3.94SDD448 pKa = 4.31YY449 pKa = 11.94NEE451 pKa = 4.51ALNGSVEE458 pKa = 4.58DD459 pKa = 4.18FWKK462 pKa = 10.24VACAIEE468 pKa = 4.5LSFF471 pKa = 5.5
Molecular weight: 51.96 kDa
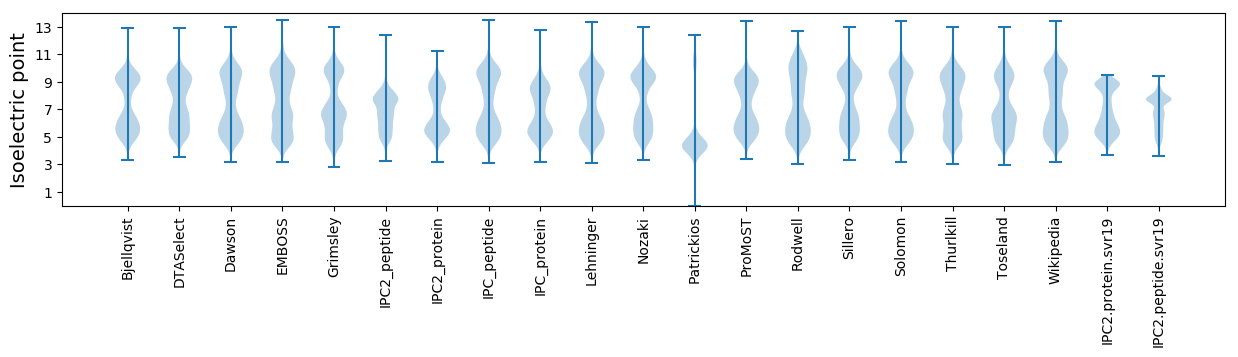
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W1ZTE0|A0A1W1ZTE0_9DELT Oxaloacetate decarboxylase gamma chain OS=Desulfobacterium vacuolatum DSM 3385 OX=1121400 GN=SAMN02746065_103163 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84IKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 9.05HH16 pKa = 5.9GFLKK20 pKa = 10.66RR21 pKa = 11.84MSTAAGRR28 pKa = 11.84RR29 pKa = 11.84IVNSRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.9GRR39 pKa = 11.84KK40 pKa = 9.09KK41 pKa = 9.67LTAA44 pKa = 4.2
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84IKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 9.05HH16 pKa = 5.9GFLKK20 pKa = 10.66RR21 pKa = 11.84MSTAAGRR28 pKa = 11.84RR29 pKa = 11.84IVNSRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.9GRR39 pKa = 11.84KK40 pKa = 9.09KK41 pKa = 9.67LTAA44 pKa = 4.2
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1418271 |
29 |
3901 |
347.4 |
38.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.836 ± 0.032 | 1.383 ± 0.016 |
5.498 ± 0.029 | 6.089 ± 0.033 |
4.561 ± 0.028 | 7.407 ± 0.033 |
2.299 ± 0.018 | 7.23 ± 0.033 |
6.538 ± 0.036 | 9.616 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.192 ± 0.019 | 4.031 ± 0.02 |
4.323 ± 0.023 | 3.316 ± 0.021 |
4.839 ± 0.027 | 5.833 ± 0.024 |
5.477 ± 0.032 | 6.661 ± 0.034 |
1.038 ± 0.014 | 2.834 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
