
Menghai rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Almendravirus; Menghai almendravirus
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
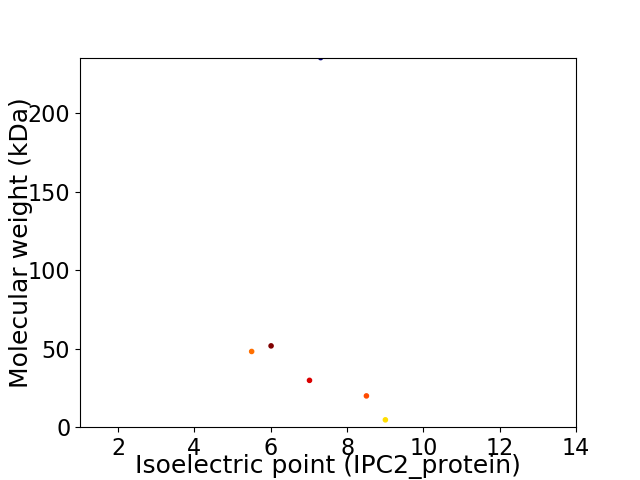
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L2YVG6|A0A1L2YVG6_9RHAB GDP polyribonucleotidyltransferase OS=Menghai rhabdovirus OX=1919071 PE=4 SV=1
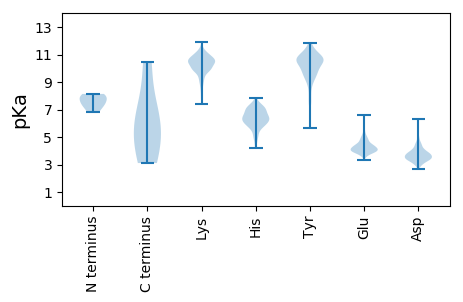
MM1 pKa = 7.98DD2 pKa = 3.66AHH4 pKa = 7.27KK5 pKa = 10.23FRR7 pKa = 11.84YY8 pKa = 9.46VDD10 pKa = 3.51GSGKK14 pKa = 10.39AWVKK18 pKa = 10.38DD19 pKa = 3.71VPSINMGDD27 pKa = 3.82SVDD30 pKa = 6.33DD31 pKa = 3.63PTTWLNDD38 pKa = 3.32GKK40 pKa = 10.52RR41 pKa = 11.84PKK43 pKa = 9.83LTIPKK48 pKa = 9.82AKK50 pKa = 10.58SNMLKK55 pKa = 9.18TVCKK59 pKa = 10.38YY60 pKa = 9.74FASQSASPEE69 pKa = 4.04NIEE72 pKa = 4.42PDD74 pKa = 3.3VALYY78 pKa = 10.72LLYY81 pKa = 10.37HH82 pKa = 6.63LSVKK86 pKa = 9.94YY87 pKa = 10.6IKK89 pKa = 10.57VDD91 pKa = 3.16IKK93 pKa = 11.5NKK95 pKa = 8.78LTLKK99 pKa = 10.72KK100 pKa = 10.39NIYY103 pKa = 9.81LNSGLKK109 pKa = 9.24TVDD112 pKa = 3.71DD113 pKa = 4.57LFDD116 pKa = 3.72VEE118 pKa = 4.92EE119 pKa = 5.12GEE121 pKa = 4.22EE122 pKa = 4.14LKK124 pKa = 11.22LSGDD128 pKa = 3.68NEE130 pKa = 4.29TSDD133 pKa = 4.49RR134 pKa = 11.84EE135 pKa = 4.18MPEE138 pKa = 4.49LEE140 pKa = 4.29ILSALLANYY149 pKa = 7.63RR150 pKa = 11.84ICGMASNADD159 pKa = 3.41QKK161 pKa = 11.37YY162 pKa = 8.49VQTLKK167 pKa = 11.12ARR169 pKa = 11.84MNRR172 pKa = 11.84TFVQKK177 pKa = 10.34QIIKK181 pKa = 10.71EE182 pKa = 4.02EE183 pKa = 4.0IQEE186 pKa = 4.06NFYY189 pKa = 11.37NALTWVNNYY198 pKa = 10.55NYY200 pKa = 9.5TRR202 pKa = 11.84LCAGIDD208 pKa = 3.47LLLFSDD214 pKa = 4.15KK215 pKa = 10.65NHH217 pKa = 7.16PYY219 pKa = 10.09SALRR223 pKa = 11.84MATQSSKK230 pKa = 10.9FKK232 pKa = 10.58DD233 pKa = 3.42CALIGEE239 pKa = 4.62INHH242 pKa = 6.59FLTTLSEE249 pKa = 4.35APMDD253 pKa = 3.69AWEE256 pKa = 4.52HH257 pKa = 5.93ALEE260 pKa = 3.98QSISKK265 pKa = 9.97EE266 pKa = 3.69IINIMTCLGRR276 pKa = 11.84GFEE279 pKa = 4.38GEE281 pKa = 4.08YY282 pKa = 11.02ANYY285 pKa = 9.26AKK287 pKa = 10.74EE288 pKa = 3.94MGAMDD293 pKa = 4.64KK294 pKa = 11.28SPFSVTANPNLHH306 pKa = 5.86NWMHH310 pKa = 6.84MICAMLSSEE319 pKa = 3.86RR320 pKa = 11.84SMNARR325 pKa = 11.84LASGNQHH332 pKa = 5.21TVFLTALACAYY343 pKa = 8.1TYY345 pKa = 10.61RR346 pKa = 11.84SKK348 pKa = 10.07TKK350 pKa = 9.58IQAVIYY356 pKa = 10.49KK357 pKa = 9.74DD358 pKa = 4.08DD359 pKa = 4.39HH360 pKa = 7.07DD361 pKa = 3.97SHH363 pKa = 7.23ARR365 pKa = 11.84SINEE369 pKa = 3.99STDD372 pKa = 3.26DD373 pKa = 3.72QIGMLEE379 pKa = 3.94TAEE382 pKa = 4.01NLKK385 pKa = 10.59ALRR388 pKa = 11.84DD389 pKa = 4.06DD390 pKa = 4.19GEE392 pKa = 4.21EE393 pKa = 4.14ASEE396 pKa = 3.49IWEE399 pKa = 3.94WFNNKK404 pKa = 7.93KK405 pKa = 9.2TIISDD410 pKa = 3.61EE411 pKa = 4.26RR412 pKa = 11.84PGTIGSYY419 pKa = 10.4LISKK423 pKa = 7.51MHH425 pKa = 6.34
MM1 pKa = 7.98DD2 pKa = 3.66AHH4 pKa = 7.27KK5 pKa = 10.23FRR7 pKa = 11.84YY8 pKa = 9.46VDD10 pKa = 3.51GSGKK14 pKa = 10.39AWVKK18 pKa = 10.38DD19 pKa = 3.71VPSINMGDD27 pKa = 3.82SVDD30 pKa = 6.33DD31 pKa = 3.63PTTWLNDD38 pKa = 3.32GKK40 pKa = 10.52RR41 pKa = 11.84PKK43 pKa = 9.83LTIPKK48 pKa = 9.82AKK50 pKa = 10.58SNMLKK55 pKa = 9.18TVCKK59 pKa = 10.38YY60 pKa = 9.74FASQSASPEE69 pKa = 4.04NIEE72 pKa = 4.42PDD74 pKa = 3.3VALYY78 pKa = 10.72LLYY81 pKa = 10.37HH82 pKa = 6.63LSVKK86 pKa = 9.94YY87 pKa = 10.6IKK89 pKa = 10.57VDD91 pKa = 3.16IKK93 pKa = 11.5NKK95 pKa = 8.78LTLKK99 pKa = 10.72KK100 pKa = 10.39NIYY103 pKa = 9.81LNSGLKK109 pKa = 9.24TVDD112 pKa = 3.71DD113 pKa = 4.57LFDD116 pKa = 3.72VEE118 pKa = 4.92EE119 pKa = 5.12GEE121 pKa = 4.22EE122 pKa = 4.14LKK124 pKa = 11.22LSGDD128 pKa = 3.68NEE130 pKa = 4.29TSDD133 pKa = 4.49RR134 pKa = 11.84EE135 pKa = 4.18MPEE138 pKa = 4.49LEE140 pKa = 4.29ILSALLANYY149 pKa = 7.63RR150 pKa = 11.84ICGMASNADD159 pKa = 3.41QKK161 pKa = 11.37YY162 pKa = 8.49VQTLKK167 pKa = 11.12ARR169 pKa = 11.84MNRR172 pKa = 11.84TFVQKK177 pKa = 10.34QIIKK181 pKa = 10.71EE182 pKa = 4.02EE183 pKa = 4.0IQEE186 pKa = 4.06NFYY189 pKa = 11.37NALTWVNNYY198 pKa = 10.55NYY200 pKa = 9.5TRR202 pKa = 11.84LCAGIDD208 pKa = 3.47LLLFSDD214 pKa = 4.15KK215 pKa = 10.65NHH217 pKa = 7.16PYY219 pKa = 10.09SALRR223 pKa = 11.84MATQSSKK230 pKa = 10.9FKK232 pKa = 10.58DD233 pKa = 3.42CALIGEE239 pKa = 4.62INHH242 pKa = 6.59FLTTLSEE249 pKa = 4.35APMDD253 pKa = 3.69AWEE256 pKa = 4.52HH257 pKa = 5.93ALEE260 pKa = 3.98QSISKK265 pKa = 9.97EE266 pKa = 3.69IINIMTCLGRR276 pKa = 11.84GFEE279 pKa = 4.38GEE281 pKa = 4.08YY282 pKa = 11.02ANYY285 pKa = 9.26AKK287 pKa = 10.74EE288 pKa = 3.94MGAMDD293 pKa = 4.64KK294 pKa = 11.28SPFSVTANPNLHH306 pKa = 5.86NWMHH310 pKa = 6.84MICAMLSSEE319 pKa = 3.86RR320 pKa = 11.84SMNARR325 pKa = 11.84LASGNQHH332 pKa = 5.21TVFLTALACAYY343 pKa = 8.1TYY345 pKa = 10.61RR346 pKa = 11.84SKK348 pKa = 10.07TKK350 pKa = 9.58IQAVIYY356 pKa = 10.49KK357 pKa = 9.74DD358 pKa = 4.08DD359 pKa = 4.39HH360 pKa = 7.07DD361 pKa = 3.97SHH363 pKa = 7.23ARR365 pKa = 11.84SINEE369 pKa = 3.99STDD372 pKa = 3.26DD373 pKa = 3.72QIGMLEE379 pKa = 3.94TAEE382 pKa = 4.01NLKK385 pKa = 10.59ALRR388 pKa = 11.84DD389 pKa = 4.06DD390 pKa = 4.19GEE392 pKa = 4.21EE393 pKa = 4.14ASEE396 pKa = 3.49IWEE399 pKa = 3.94WFNNKK404 pKa = 7.93KK405 pKa = 9.2TIISDD410 pKa = 3.61EE411 pKa = 4.26RR412 pKa = 11.84PGTIGSYY419 pKa = 10.4LISKK423 pKa = 7.51MHH425 pKa = 6.34
Molecular weight: 48.22 kDa
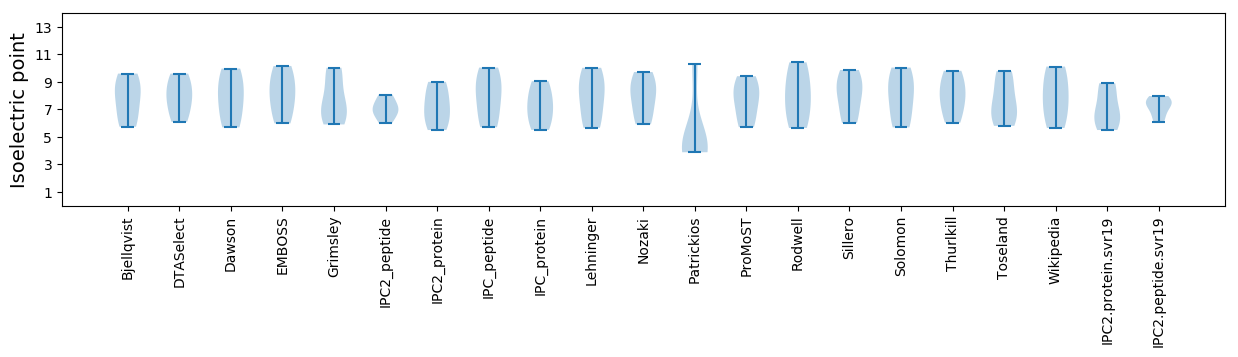
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L2YVG9|A0A1L2YVG9_9RHAB G protein OS=Menghai rhabdovirus OX=1919071 PE=4 SV=1
MM1 pKa = 7.35TKK3 pKa = 10.4NIRR6 pKa = 11.84TILSDD11 pKa = 4.08FILRR15 pKa = 11.84QKK17 pKa = 9.38IAKK20 pKa = 10.02DD21 pKa = 2.74IGEE24 pKa = 4.4DD25 pKa = 3.13KK26 pKa = 11.05KK27 pKa = 11.08GFYY30 pKa = 10.15YY31 pKa = 10.73SRR33 pKa = 11.84IDD35 pKa = 3.72GPIMNN40 pKa = 5.24
MM1 pKa = 7.35TKK3 pKa = 10.4NIRR6 pKa = 11.84TILSDD11 pKa = 4.08FILRR15 pKa = 11.84QKK17 pKa = 9.38IAKK20 pKa = 10.02DD21 pKa = 2.74IGEE24 pKa = 4.4DD25 pKa = 3.13KK26 pKa = 11.05KK27 pKa = 11.08GFYY30 pKa = 10.15YY31 pKa = 10.73SRR33 pKa = 11.84IDD35 pKa = 3.72GPIMNN40 pKa = 5.24
Molecular weight: 4.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3364 |
40 |
2032 |
560.7 |
64.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.805 ± 0.88 | 1.486 ± 0.368 |
6.361 ± 0.389 | 6.897 ± 0.633 |
4.102 ± 0.395 | 4.251 ± 0.238 |
2.14 ± 0.217 | 8.383 ± 0.678 |
9.721 ± 1.343 | 9.245 ± 0.947 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.032 ± 0.367 | 7.134 ± 0.191 |
3.151 ± 0.103 | 2.497 ± 0.302 |
3.686 ± 0.531 | 8.205 ± 0.764 |
4.637 ± 0.491 | 4.786 ± 0.388 |
1.249 ± 0.119 | 5.232 ± 0.482 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
