
Ralstonia phage Simangalove
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Bakolyvirus; Ralstonia virus Simangalove
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
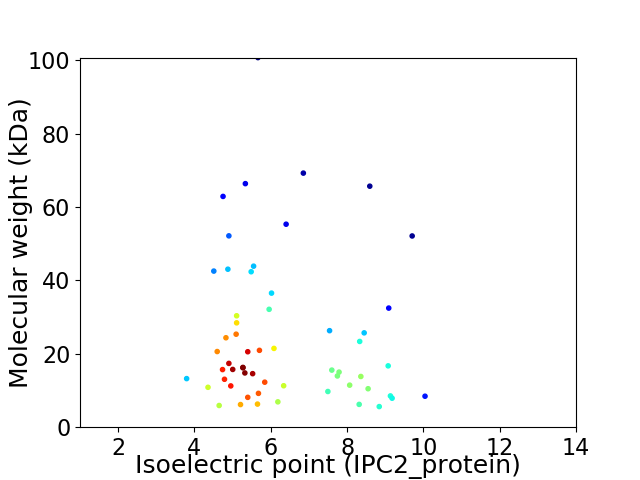
Virtual 2D-PAGE plot for 58 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
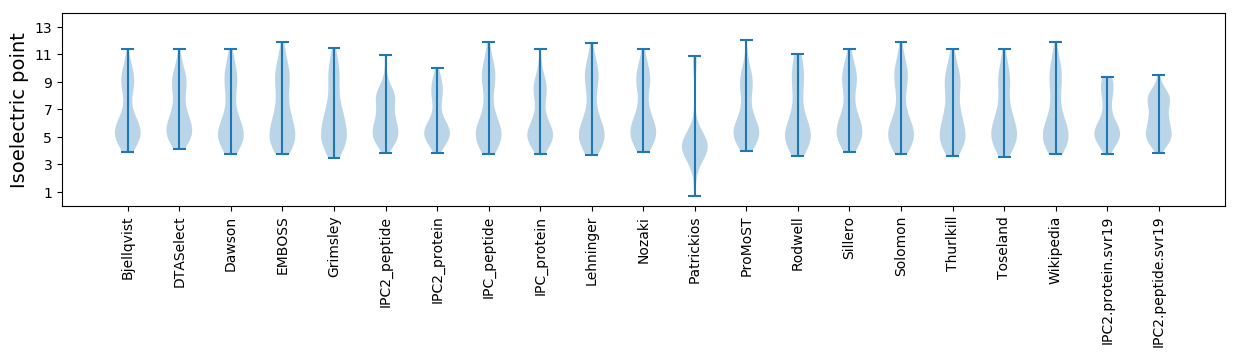
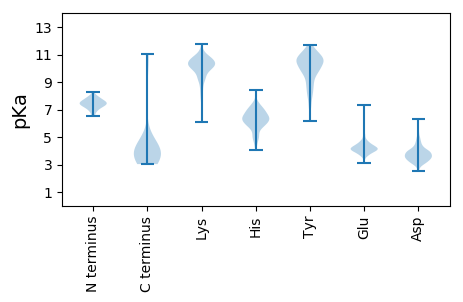
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7G5BBR4|A0A7G5BBR4_9CAUD Uncharacterized protein OS=Ralstonia phage Simangalove OX=2759710 GN=R1_00035 PE=4 SV=1
MM1 pKa = 7.82DD2 pKa = 4.71TLLLDD7 pKa = 3.81TSTWDD12 pKa = 3.36LTTDD16 pKa = 3.19ASGNLATVGDD26 pKa = 4.47ADD28 pKa = 4.71RR29 pKa = 11.84DD30 pKa = 3.98ATNGGALRR38 pKa = 11.84LAQDD42 pKa = 3.25VAARR46 pKa = 11.84VSSWRR51 pKa = 11.84GEE53 pKa = 4.1VYY55 pKa = 10.89YY56 pKa = 9.65DD57 pKa = 3.35TTQGIPYY64 pKa = 8.49DD65 pKa = 4.38TILGEE70 pKa = 4.18YY71 pKa = 8.8PNLVVLQQLYY81 pKa = 10.57SEE83 pKa = 4.84EE84 pKa = 4.2SLNVPGCATALAEE97 pKa = 4.16LTYY100 pKa = 11.24DD101 pKa = 3.22RR102 pKa = 11.84AGRR105 pKa = 11.84LLGGTINVSDD115 pKa = 3.62ISGNPAVVTLL125 pKa = 4.35
MM1 pKa = 7.82DD2 pKa = 4.71TLLLDD7 pKa = 3.81TSTWDD12 pKa = 3.36LTTDD16 pKa = 3.19ASGNLATVGDD26 pKa = 4.47ADD28 pKa = 4.71RR29 pKa = 11.84DD30 pKa = 3.98ATNGGALRR38 pKa = 11.84LAQDD42 pKa = 3.25VAARR46 pKa = 11.84VSSWRR51 pKa = 11.84GEE53 pKa = 4.1VYY55 pKa = 10.89YY56 pKa = 9.65DD57 pKa = 3.35TTQGIPYY64 pKa = 8.49DD65 pKa = 4.38TILGEE70 pKa = 4.18YY71 pKa = 8.8PNLVVLQQLYY81 pKa = 10.57SEE83 pKa = 4.84EE84 pKa = 4.2SLNVPGCATALAEE97 pKa = 4.16LTYY100 pKa = 11.24DD101 pKa = 3.22RR102 pKa = 11.84AGRR105 pKa = 11.84LLGGTINVSDD115 pKa = 3.62ISGNPAVVTLL125 pKa = 4.35
Molecular weight: 13.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7G5BBS8|A0A7G5BBS8_9CAUD Uncharacterized protein OS=Ralstonia phage Simangalove OX=2759710 GN=R1_00049 PE=4 SV=1
MM1 pKa = 7.31SVFVLGRR8 pKa = 11.84NGAPLMPCSEE18 pKa = 3.67QRR20 pKa = 11.84ARR22 pKa = 11.84KK23 pKa = 9.03LLSAGRR29 pKa = 11.84ARR31 pKa = 11.84VHH33 pKa = 6.32RR34 pKa = 11.84LYY36 pKa = 10.32PFTIRR41 pKa = 11.84LIDD44 pKa = 3.61RR45 pKa = 11.84QQANSVLQPLEE56 pKa = 4.54LKK58 pKa = 10.42LDD60 pKa = 4.35PGSKK64 pKa = 6.87TTGLAVCRR72 pKa = 11.84VEE74 pKa = 4.85EE75 pKa = 4.68RR76 pKa = 11.84IDD78 pKa = 3.29VDD80 pKa = 3.97GVVEE84 pKa = 4.1RR85 pKa = 11.84VMHH88 pKa = 6.39IRR90 pKa = 11.84FLMDD94 pKa = 3.92LVHH97 pKa = 6.97RR98 pKa = 11.84GHH100 pKa = 7.53AIKK103 pKa = 10.34EE104 pKa = 4.13ALHH107 pKa = 6.32ARR109 pKa = 11.84AAMRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84GNLRR121 pKa = 11.84YY122 pKa = 9.46RR123 pKa = 11.84APRR126 pKa = 11.84FDD128 pKa = 2.98NRR130 pKa = 11.84TRR132 pKa = 11.84KK133 pKa = 9.91EE134 pKa = 3.62GWLPPSLQHH143 pKa = 7.23RR144 pKa = 11.84VDD146 pKa = 3.81TTVSWVKK153 pKa = 10.15RR154 pKa = 11.84LCRR157 pKa = 11.84LAPITHH163 pKa = 7.53LAQEE167 pKa = 4.45LVRR170 pKa = 11.84FDD172 pKa = 3.56MQLMQNAEE180 pKa = 3.87ISGVEE185 pKa = 4.02YY186 pKa = 10.62QQGEE190 pKa = 4.04LAGYY194 pKa = 7.23EE195 pKa = 3.88VRR197 pKa = 11.84EE198 pKa = 3.92YY199 pKa = 11.21LLEE202 pKa = 4.35KK203 pKa = 10.41FGRR206 pKa = 11.84TCQYY210 pKa = 10.63CDD212 pKa = 3.25EE213 pKa = 4.8TGVPLQVEE221 pKa = 4.92HH222 pKa = 6.68IHH224 pKa = 6.08PKK226 pKa = 10.4ARR228 pKa = 11.84GGSNRR233 pKa = 11.84VSNLTLACQPCNEE246 pKa = 4.9KK247 pKa = 10.41KK248 pKa = 10.31DD249 pKa = 3.8SQDD252 pKa = 2.61IRR254 pKa = 11.84VFLAKK259 pKa = 10.57DD260 pKa = 3.18PMRR263 pKa = 11.84LEE265 pKa = 5.13RR266 pKa = 11.84ILARR270 pKa = 11.84AKK272 pKa = 10.84APLRR276 pKa = 11.84DD277 pKa = 3.54AAAVNATRR285 pKa = 11.84WALLNALKK293 pKa = 9.42GTGLPVTTGSGGQTKK308 pKa = 9.43WNRR311 pKa = 11.84TRR313 pKa = 11.84LGLPKK318 pKa = 9.65THH320 pKa = 6.85SLDD323 pKa = 3.5AACVGRR329 pKa = 11.84ADD331 pKa = 3.88RR332 pKa = 11.84VTGSGAPVLVVKK344 pKa = 9.21CTGRR348 pKa = 11.84GSRR351 pKa = 11.84SRR353 pKa = 11.84TRR355 pKa = 11.84LNQYY359 pKa = 8.61GFPRR363 pKa = 11.84AYY365 pKa = 10.63LARR368 pKa = 11.84NKK370 pKa = 9.22TAFGFRR376 pKa = 11.84TGDD379 pKa = 3.32MVIASVPSGKK389 pKa = 10.13KK390 pKa = 9.39AGTYY394 pKa = 9.16KK395 pKa = 10.63GRR397 pKa = 11.84VAIRR401 pKa = 11.84QTGSFNIQPGKK412 pKa = 9.8PGLPTVQGISHH423 pKa = 5.92KK424 pKa = 9.82HH425 pKa = 5.11CRR427 pKa = 11.84VAQRR431 pKa = 11.84GDD433 pKa = 3.13GYY435 pKa = 11.29GYY437 pKa = 11.49ALASPANSQPGKK449 pKa = 9.96AKK451 pKa = 10.4RR452 pKa = 11.84PAASALYY459 pKa = 10.63LLGLKK464 pKa = 10.53AEE466 pKa = 4.36VSLADD471 pKa = 3.42
MM1 pKa = 7.31SVFVLGRR8 pKa = 11.84NGAPLMPCSEE18 pKa = 3.67QRR20 pKa = 11.84ARR22 pKa = 11.84KK23 pKa = 9.03LLSAGRR29 pKa = 11.84ARR31 pKa = 11.84VHH33 pKa = 6.32RR34 pKa = 11.84LYY36 pKa = 10.32PFTIRR41 pKa = 11.84LIDD44 pKa = 3.61RR45 pKa = 11.84QQANSVLQPLEE56 pKa = 4.54LKK58 pKa = 10.42LDD60 pKa = 4.35PGSKK64 pKa = 6.87TTGLAVCRR72 pKa = 11.84VEE74 pKa = 4.85EE75 pKa = 4.68RR76 pKa = 11.84IDD78 pKa = 3.29VDD80 pKa = 3.97GVVEE84 pKa = 4.1RR85 pKa = 11.84VMHH88 pKa = 6.39IRR90 pKa = 11.84FLMDD94 pKa = 3.92LVHH97 pKa = 6.97RR98 pKa = 11.84GHH100 pKa = 7.53AIKK103 pKa = 10.34EE104 pKa = 4.13ALHH107 pKa = 6.32ARR109 pKa = 11.84AAMRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84GNLRR121 pKa = 11.84YY122 pKa = 9.46RR123 pKa = 11.84APRR126 pKa = 11.84FDD128 pKa = 2.98NRR130 pKa = 11.84TRR132 pKa = 11.84KK133 pKa = 9.91EE134 pKa = 3.62GWLPPSLQHH143 pKa = 7.23RR144 pKa = 11.84VDD146 pKa = 3.81TTVSWVKK153 pKa = 10.15RR154 pKa = 11.84LCRR157 pKa = 11.84LAPITHH163 pKa = 7.53LAQEE167 pKa = 4.45LVRR170 pKa = 11.84FDD172 pKa = 3.56MQLMQNAEE180 pKa = 3.87ISGVEE185 pKa = 4.02YY186 pKa = 10.62QQGEE190 pKa = 4.04LAGYY194 pKa = 7.23EE195 pKa = 3.88VRR197 pKa = 11.84EE198 pKa = 3.92YY199 pKa = 11.21LLEE202 pKa = 4.35KK203 pKa = 10.41FGRR206 pKa = 11.84TCQYY210 pKa = 10.63CDD212 pKa = 3.25EE213 pKa = 4.8TGVPLQVEE221 pKa = 4.92HH222 pKa = 6.68IHH224 pKa = 6.08PKK226 pKa = 10.4ARR228 pKa = 11.84GGSNRR233 pKa = 11.84VSNLTLACQPCNEE246 pKa = 4.9KK247 pKa = 10.41KK248 pKa = 10.31DD249 pKa = 3.8SQDD252 pKa = 2.61IRR254 pKa = 11.84VFLAKK259 pKa = 10.57DD260 pKa = 3.18PMRR263 pKa = 11.84LEE265 pKa = 5.13RR266 pKa = 11.84ILARR270 pKa = 11.84AKK272 pKa = 10.84APLRR276 pKa = 11.84DD277 pKa = 3.54AAAVNATRR285 pKa = 11.84WALLNALKK293 pKa = 9.42GTGLPVTTGSGGQTKK308 pKa = 9.43WNRR311 pKa = 11.84TRR313 pKa = 11.84LGLPKK318 pKa = 9.65THH320 pKa = 6.85SLDD323 pKa = 3.5AACVGRR329 pKa = 11.84ADD331 pKa = 3.88RR332 pKa = 11.84VTGSGAPVLVVKK344 pKa = 9.21CTGRR348 pKa = 11.84GSRR351 pKa = 11.84SRR353 pKa = 11.84TRR355 pKa = 11.84LNQYY359 pKa = 8.61GFPRR363 pKa = 11.84AYY365 pKa = 10.63LARR368 pKa = 11.84NKK370 pKa = 9.22TAFGFRR376 pKa = 11.84TGDD379 pKa = 3.32MVIASVPSGKK389 pKa = 10.13KK390 pKa = 9.39AGTYY394 pKa = 9.16KK395 pKa = 10.63GRR397 pKa = 11.84VAIRR401 pKa = 11.84QTGSFNIQPGKK412 pKa = 9.8PGLPTVQGISHH423 pKa = 5.92KK424 pKa = 9.82HH425 pKa = 5.11CRR427 pKa = 11.84VAQRR431 pKa = 11.84GDD433 pKa = 3.13GYY435 pKa = 11.29GYY437 pKa = 11.49ALASPANSQPGKK449 pKa = 9.96AKK451 pKa = 10.4RR452 pKa = 11.84PAASALYY459 pKa = 10.63LLGLKK464 pKa = 10.53AEE466 pKa = 4.36VSLADD471 pKa = 3.42
Molecular weight: 52.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13237 |
52 |
917 |
228.2 |
24.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.979 ± 0.428 | 0.997 ± 0.114 |
5.636 ± 0.257 | 5.092 ± 0.375 |
3.294 ± 0.202 | 8.038 ± 0.581 |
1.76 ± 0.178 | 4.132 ± 0.172 |
3.611 ± 0.284 | 8.393 ± 0.32 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.251 ± 0.156 | 3.709 ± 0.23 |
5.613 ± 0.229 | 4.314 ± 0.189 |
6.285 ± 0.42 | 5.651 ± 0.317 |
6.716 ± 0.444 | 7.147 ± 0.279 |
1.518 ± 0.127 | 2.863 ± 0.178 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
