
Wenzhou crab virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
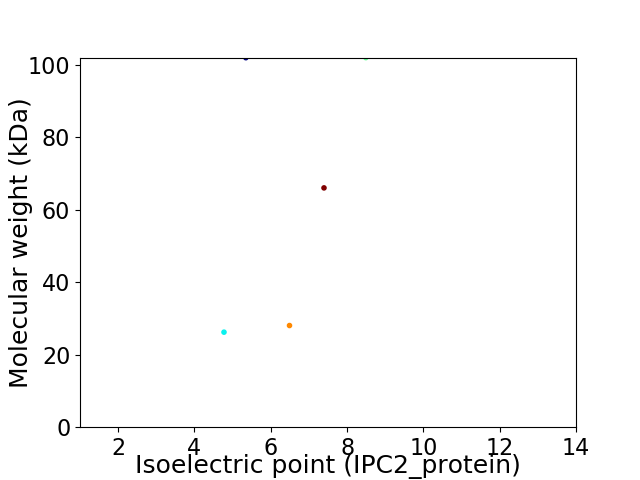
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFJ5|A0A1L3KFJ5_9VIRU RdRP_1 domain-containing protein OS=Wenzhou crab virus 5 OX=1923562 PE=4 SV=1
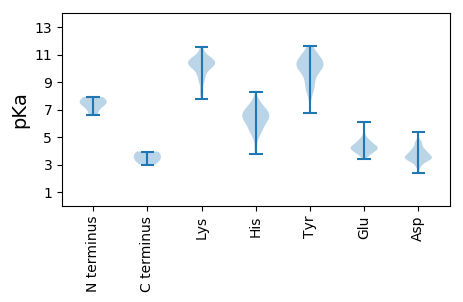
MM1 pKa = 7.52LARR4 pKa = 11.84ARR6 pKa = 11.84MMTPQKK12 pKa = 10.6LAVCLPVGQSLYY24 pKa = 10.56GWYY27 pKa = 10.63SSMLAVSWGTHH38 pKa = 4.84LRR40 pKa = 11.84RR41 pKa = 11.84WPQNSGGPPFYY52 pKa = 10.6LRR54 pKa = 11.84TMTLEE59 pKa = 4.26PSSAIYY65 pKa = 9.88TMSRR69 pKa = 11.84YY70 pKa = 8.4FVCSSDD76 pKa = 3.75EE77 pKa = 4.21EE78 pKa = 4.39EE79 pKa = 5.37LNEE82 pKa = 4.63QYY84 pKa = 10.94EE85 pKa = 4.28LQLAGSSQSDD95 pKa = 3.45ANSCAEE101 pKa = 3.69GSTAGFCGLQSVSAQSVPDD120 pKa = 3.62VDD122 pKa = 4.18SSEE125 pKa = 4.13PACVCRR131 pKa = 11.84CFSTPHH137 pKa = 6.73GPTCYY142 pKa = 8.85VTRR145 pKa = 11.84GGVRR149 pKa = 11.84ALVEE153 pKa = 3.86ATAGIFSEE161 pKa = 4.23HH162 pKa = 6.32TGRR165 pKa = 11.84STRR168 pKa = 11.84TRR170 pKa = 11.84VILGALGATVLGAAAGAVTYY190 pKa = 10.65YY191 pKa = 11.26SLTTSGGTDD200 pKa = 3.05SLSEE204 pKa = 4.2SGRR207 pKa = 11.84EE208 pKa = 3.96ALGDD212 pKa = 3.52AVAQVSLNDD221 pKa = 3.75LPIVHH226 pKa = 7.35PEE228 pKa = 3.96STSPPSADD236 pKa = 3.08GSEE239 pKa = 3.96VSAWDD244 pKa = 3.62NQYY247 pKa = 11.51DD248 pKa = 3.41
MM1 pKa = 7.52LARR4 pKa = 11.84ARR6 pKa = 11.84MMTPQKK12 pKa = 10.6LAVCLPVGQSLYY24 pKa = 10.56GWYY27 pKa = 10.63SSMLAVSWGTHH38 pKa = 4.84LRR40 pKa = 11.84RR41 pKa = 11.84WPQNSGGPPFYY52 pKa = 10.6LRR54 pKa = 11.84TMTLEE59 pKa = 4.26PSSAIYY65 pKa = 9.88TMSRR69 pKa = 11.84YY70 pKa = 8.4FVCSSDD76 pKa = 3.75EE77 pKa = 4.21EE78 pKa = 4.39EE79 pKa = 5.37LNEE82 pKa = 4.63QYY84 pKa = 10.94EE85 pKa = 4.28LQLAGSSQSDD95 pKa = 3.45ANSCAEE101 pKa = 3.69GSTAGFCGLQSVSAQSVPDD120 pKa = 3.62VDD122 pKa = 4.18SSEE125 pKa = 4.13PACVCRR131 pKa = 11.84CFSTPHH137 pKa = 6.73GPTCYY142 pKa = 8.85VTRR145 pKa = 11.84GGVRR149 pKa = 11.84ALVEE153 pKa = 3.86ATAGIFSEE161 pKa = 4.23HH162 pKa = 6.32TGRR165 pKa = 11.84STRR168 pKa = 11.84TRR170 pKa = 11.84VILGALGATVLGAAAGAVTYY190 pKa = 10.65YY191 pKa = 11.26SLTTSGGTDD200 pKa = 3.05SLSEE204 pKa = 4.2SGRR207 pKa = 11.84EE208 pKa = 3.96ALGDD212 pKa = 3.52AVAQVSLNDD221 pKa = 3.75LPIVHH226 pKa = 7.35PEE228 pKa = 3.96STSPPSADD236 pKa = 3.08GSEE239 pKa = 3.96VSAWDD244 pKa = 3.62NQYY247 pKa = 11.51DD248 pKa = 3.41
Molecular weight: 26.2 kDa
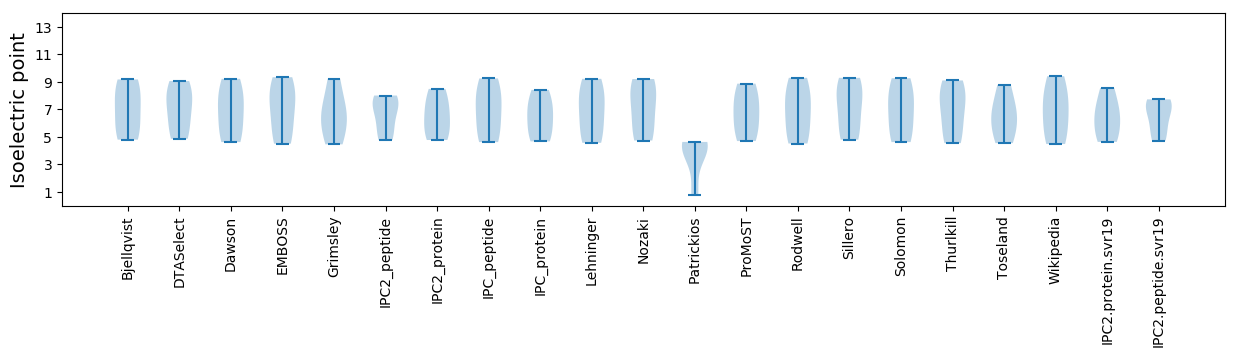
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFJ5|A0A1L3KFJ5_9VIRU RdRP_1 domain-containing protein OS=Wenzhou crab virus 5 OX=1923562 PE=4 SV=1
MM1 pKa = 6.57YY2 pKa = 9.73TPGLSLGTTLARR14 pKa = 11.84LALSGPRR21 pKa = 11.84MQPIPNHH28 pKa = 5.61MSGDD32 pKa = 3.97EE33 pKa = 4.29DD34 pKa = 3.83SLSQLFEE41 pKa = 4.0RR42 pKa = 11.84LKK44 pKa = 11.26LIVTQPALIPEE55 pKa = 4.64LAGDD59 pKa = 3.81YY60 pKa = 10.82VPAGVQQLFEE70 pKa = 4.22QMRR73 pKa = 11.84GDD75 pKa = 3.34AALPRR80 pKa = 11.84WFYY83 pKa = 11.25QHH85 pKa = 6.85LSAPRR90 pKa = 11.84ALSLRR95 pKa = 11.84PGTQRR100 pKa = 11.84PTCQGMDD107 pKa = 4.1RR108 pKa = 11.84ITGLPEE114 pKa = 4.89AGGLPIPVFRR124 pKa = 11.84AVAEE128 pKa = 4.51EE129 pKa = 3.56IRR131 pKa = 11.84AEE133 pKa = 3.91RR134 pKa = 11.84RR135 pKa = 11.84QVGTDD140 pKa = 3.27PRR142 pKa = 11.84AVWFQEE148 pKa = 3.37KK149 pKa = 10.11HH150 pKa = 5.92RR151 pKa = 11.84LHH153 pKa = 6.99LPLTKK158 pKa = 10.45DD159 pKa = 2.89EE160 pKa = 5.08LGRR163 pKa = 11.84QRR165 pKa = 11.84YY166 pKa = 8.9IPTFRR171 pKa = 11.84WKK173 pKa = 10.14PDD175 pKa = 3.01VFGPRR180 pKa = 11.84FSLGIGMYY188 pKa = 10.45YY189 pKa = 10.31EE190 pKa = 4.38VLSHH194 pKa = 5.91EE195 pKa = 4.36QRR197 pKa = 11.84STLTSWLEE205 pKa = 3.66RR206 pKa = 11.84LEE208 pKa = 4.42PYY210 pKa = 9.96IALCQSTLTVVAIFKK225 pKa = 10.58AVEE228 pKa = 4.05GWAKK232 pKa = 10.61AAGDD236 pKa = 4.05LFKK239 pKa = 10.49HH240 pKa = 5.28WRR242 pKa = 11.84IPCGIALLGGYY253 pKa = 9.15QPAPPLVFFRR263 pKa = 11.84DD264 pKa = 4.03EE265 pKa = 3.91VSEE268 pKa = 4.28WVSGPLPDD276 pKa = 5.14IGQHH280 pKa = 5.26LPGWRR285 pKa = 11.84DD286 pKa = 3.27SFEE289 pKa = 4.14WASKK293 pKa = 10.59QLLTGCGPEE302 pKa = 3.97QPPDD306 pKa = 3.41TFEE309 pKa = 6.01DD310 pKa = 4.33YY311 pKa = 11.08VLRR314 pKa = 11.84GDD316 pKa = 3.7WVRR319 pKa = 11.84PGATDD324 pKa = 2.88QGTFPSVTSRR334 pKa = 11.84GEE336 pKa = 3.76IVSVRR341 pKa = 11.84KK342 pKa = 8.18TKK344 pKa = 10.77VAMFAATTPDD354 pKa = 3.82DD355 pKa = 4.68LLALAYY361 pKa = 10.65SPAQPPRR368 pKa = 11.84AMVKK372 pKa = 10.3RR373 pKa = 11.84EE374 pKa = 3.97LTKK377 pKa = 10.82ARR379 pKa = 11.84AVVITDD385 pKa = 3.38TPSYY389 pKa = 11.24LRR391 pKa = 11.84MSWISTALEE400 pKa = 3.72PLLRR404 pKa = 11.84RR405 pKa = 11.84SRR407 pKa = 11.84NASVFFSGKK416 pKa = 9.68RR417 pKa = 11.84LAKK420 pKa = 9.09MWADD424 pKa = 3.87LASDD428 pKa = 3.91SADD431 pKa = 3.2VTLWKK436 pKa = 10.77CPIDD440 pKa = 3.75QSKK443 pKa = 9.48FDD445 pKa = 4.09HH446 pKa = 6.32MVEE449 pKa = 4.22PWALSVILDD458 pKa = 4.4RR459 pKa = 11.84IEE461 pKa = 3.77EE462 pKa = 4.85LIMGDD467 pKa = 3.45QAKK470 pKa = 10.05HH471 pKa = 4.13VLRR474 pKa = 11.84LIRR477 pKa = 11.84RR478 pKa = 11.84DD479 pKa = 4.17LIPEE483 pKa = 4.32KK484 pKa = 10.55GTVTVGDD491 pKa = 3.67ADD493 pKa = 3.91VPVTKK498 pKa = 10.37GVLSGWRR505 pKa = 11.84WTALLDD511 pKa = 3.22TWVNFSQVVGSLHH524 pKa = 5.38YY525 pKa = 10.31LYY527 pKa = 10.46TRR529 pKa = 11.84GVVPSIDD536 pKa = 3.31YY537 pKa = 9.06TGWYY541 pKa = 8.94FQGDD545 pKa = 3.8DD546 pKa = 3.27ARR548 pKa = 11.84LVVRR552 pKa = 11.84NPGIAVAVAEE562 pKa = 4.39VMRR565 pKa = 11.84EE566 pKa = 4.01AGLKK570 pKa = 9.18IHH572 pKa = 7.17PSKK575 pKa = 9.81TWLSPFRR582 pKa = 11.84DD583 pKa = 3.29EE584 pKa = 5.52FLRR587 pKa = 11.84QVAEE591 pKa = 4.63DD592 pKa = 4.06NVTMGYY598 pKa = 10.01LARR601 pKa = 11.84SVPALLWRR609 pKa = 11.84NPINAAADD617 pKa = 3.5NWPAAARR624 pKa = 11.84AAVGRR629 pKa = 11.84WDD631 pKa = 3.58GLVLRR636 pKa = 11.84GADD639 pKa = 3.44PYY641 pKa = 11.08RR642 pKa = 11.84VLEE645 pKa = 4.24HH646 pKa = 6.72ALKK649 pKa = 10.42EE650 pKa = 4.12LTHH653 pKa = 6.92LIPYY657 pKa = 8.74QRR659 pKa = 11.84STIHH663 pKa = 6.3KK664 pKa = 8.35WLSLPPHH671 pKa = 6.74LGGAGVTWVTPDD683 pKa = 3.52LTSLALIRR691 pKa = 11.84YY692 pKa = 6.98PLEE695 pKa = 4.5RR696 pKa = 11.84SLLRR700 pKa = 11.84FDD702 pKa = 4.59GVGLFDD708 pKa = 4.37RR709 pKa = 11.84LVSQVDD715 pKa = 3.87LPRR718 pKa = 11.84DD719 pKa = 3.34EE720 pKa = 6.1LYY722 pKa = 10.96AKK724 pKa = 10.06CYY726 pKa = 10.13QYY728 pKa = 11.51LSDD731 pKa = 4.59VVDD734 pKa = 3.45THH736 pKa = 6.18EE737 pKa = 4.79PKK739 pKa = 10.4HH740 pKa = 6.18LYY742 pKa = 10.08RR743 pKa = 11.84DD744 pKa = 3.55ILGSMSKK751 pKa = 8.35PTLDD755 pKa = 2.62WGVPPKK761 pKa = 10.0PWPPLGLVPCGAPPAQPTWARR782 pKa = 11.84GLQFPNWFRR791 pKa = 11.84NIAIDD796 pKa = 3.47VAVRR800 pKa = 11.84TKK802 pKa = 9.01TEE804 pKa = 3.58KK805 pKa = 10.83SVDD808 pKa = 3.66VLFEE812 pKa = 4.71PYY814 pKa = 9.74SASHH818 pKa = 6.01SPIIRR823 pKa = 11.84ARR825 pKa = 11.84HH826 pKa = 5.29GRR828 pKa = 11.84NTWADD833 pKa = 3.3WAKK836 pKa = 10.73GALSLPVRR844 pKa = 11.84CWPGWDD850 pKa = 3.21KK851 pKa = 11.5GYY853 pKa = 10.63LATQYY858 pKa = 10.52RR859 pKa = 11.84ARR861 pKa = 11.84IDD863 pKa = 3.58TFVRR867 pKa = 11.84CYY869 pKa = 10.37LGSSRR874 pKa = 11.84RR875 pKa = 11.84WGSRR879 pKa = 11.84ARR881 pKa = 11.84LASEE885 pKa = 4.01VFFYY889 pKa = 10.79SIQRR893 pKa = 11.84CPDD896 pKa = 2.69SLYY899 pKa = 10.05PYY901 pKa = 10.08RR902 pKa = 11.84IAAA905 pKa = 3.95
MM1 pKa = 6.57YY2 pKa = 9.73TPGLSLGTTLARR14 pKa = 11.84LALSGPRR21 pKa = 11.84MQPIPNHH28 pKa = 5.61MSGDD32 pKa = 3.97EE33 pKa = 4.29DD34 pKa = 3.83SLSQLFEE41 pKa = 4.0RR42 pKa = 11.84LKK44 pKa = 11.26LIVTQPALIPEE55 pKa = 4.64LAGDD59 pKa = 3.81YY60 pKa = 10.82VPAGVQQLFEE70 pKa = 4.22QMRR73 pKa = 11.84GDD75 pKa = 3.34AALPRR80 pKa = 11.84WFYY83 pKa = 11.25QHH85 pKa = 6.85LSAPRR90 pKa = 11.84ALSLRR95 pKa = 11.84PGTQRR100 pKa = 11.84PTCQGMDD107 pKa = 4.1RR108 pKa = 11.84ITGLPEE114 pKa = 4.89AGGLPIPVFRR124 pKa = 11.84AVAEE128 pKa = 4.51EE129 pKa = 3.56IRR131 pKa = 11.84AEE133 pKa = 3.91RR134 pKa = 11.84RR135 pKa = 11.84QVGTDD140 pKa = 3.27PRR142 pKa = 11.84AVWFQEE148 pKa = 3.37KK149 pKa = 10.11HH150 pKa = 5.92RR151 pKa = 11.84LHH153 pKa = 6.99LPLTKK158 pKa = 10.45DD159 pKa = 2.89EE160 pKa = 5.08LGRR163 pKa = 11.84QRR165 pKa = 11.84YY166 pKa = 8.9IPTFRR171 pKa = 11.84WKK173 pKa = 10.14PDD175 pKa = 3.01VFGPRR180 pKa = 11.84FSLGIGMYY188 pKa = 10.45YY189 pKa = 10.31EE190 pKa = 4.38VLSHH194 pKa = 5.91EE195 pKa = 4.36QRR197 pKa = 11.84STLTSWLEE205 pKa = 3.66RR206 pKa = 11.84LEE208 pKa = 4.42PYY210 pKa = 9.96IALCQSTLTVVAIFKK225 pKa = 10.58AVEE228 pKa = 4.05GWAKK232 pKa = 10.61AAGDD236 pKa = 4.05LFKK239 pKa = 10.49HH240 pKa = 5.28WRR242 pKa = 11.84IPCGIALLGGYY253 pKa = 9.15QPAPPLVFFRR263 pKa = 11.84DD264 pKa = 4.03EE265 pKa = 3.91VSEE268 pKa = 4.28WVSGPLPDD276 pKa = 5.14IGQHH280 pKa = 5.26LPGWRR285 pKa = 11.84DD286 pKa = 3.27SFEE289 pKa = 4.14WASKK293 pKa = 10.59QLLTGCGPEE302 pKa = 3.97QPPDD306 pKa = 3.41TFEE309 pKa = 6.01DD310 pKa = 4.33YY311 pKa = 11.08VLRR314 pKa = 11.84GDD316 pKa = 3.7WVRR319 pKa = 11.84PGATDD324 pKa = 2.88QGTFPSVTSRR334 pKa = 11.84GEE336 pKa = 3.76IVSVRR341 pKa = 11.84KK342 pKa = 8.18TKK344 pKa = 10.77VAMFAATTPDD354 pKa = 3.82DD355 pKa = 4.68LLALAYY361 pKa = 10.65SPAQPPRR368 pKa = 11.84AMVKK372 pKa = 10.3RR373 pKa = 11.84EE374 pKa = 3.97LTKK377 pKa = 10.82ARR379 pKa = 11.84AVVITDD385 pKa = 3.38TPSYY389 pKa = 11.24LRR391 pKa = 11.84MSWISTALEE400 pKa = 3.72PLLRR404 pKa = 11.84RR405 pKa = 11.84SRR407 pKa = 11.84NASVFFSGKK416 pKa = 9.68RR417 pKa = 11.84LAKK420 pKa = 9.09MWADD424 pKa = 3.87LASDD428 pKa = 3.91SADD431 pKa = 3.2VTLWKK436 pKa = 10.77CPIDD440 pKa = 3.75QSKK443 pKa = 9.48FDD445 pKa = 4.09HH446 pKa = 6.32MVEE449 pKa = 4.22PWALSVILDD458 pKa = 4.4RR459 pKa = 11.84IEE461 pKa = 3.77EE462 pKa = 4.85LIMGDD467 pKa = 3.45QAKK470 pKa = 10.05HH471 pKa = 4.13VLRR474 pKa = 11.84LIRR477 pKa = 11.84RR478 pKa = 11.84DD479 pKa = 4.17LIPEE483 pKa = 4.32KK484 pKa = 10.55GTVTVGDD491 pKa = 3.67ADD493 pKa = 3.91VPVTKK498 pKa = 10.37GVLSGWRR505 pKa = 11.84WTALLDD511 pKa = 3.22TWVNFSQVVGSLHH524 pKa = 5.38YY525 pKa = 10.31LYY527 pKa = 10.46TRR529 pKa = 11.84GVVPSIDD536 pKa = 3.31YY537 pKa = 9.06TGWYY541 pKa = 8.94FQGDD545 pKa = 3.8DD546 pKa = 3.27ARR548 pKa = 11.84LVVRR552 pKa = 11.84NPGIAVAVAEE562 pKa = 4.39VMRR565 pKa = 11.84EE566 pKa = 4.01AGLKK570 pKa = 9.18IHH572 pKa = 7.17PSKK575 pKa = 9.81TWLSPFRR582 pKa = 11.84DD583 pKa = 3.29EE584 pKa = 5.52FLRR587 pKa = 11.84QVAEE591 pKa = 4.63DD592 pKa = 4.06NVTMGYY598 pKa = 10.01LARR601 pKa = 11.84SVPALLWRR609 pKa = 11.84NPINAAADD617 pKa = 3.5NWPAAARR624 pKa = 11.84AAVGRR629 pKa = 11.84WDD631 pKa = 3.58GLVLRR636 pKa = 11.84GADD639 pKa = 3.44PYY641 pKa = 11.08RR642 pKa = 11.84VLEE645 pKa = 4.24HH646 pKa = 6.72ALKK649 pKa = 10.42EE650 pKa = 4.12LTHH653 pKa = 6.92LIPYY657 pKa = 8.74QRR659 pKa = 11.84STIHH663 pKa = 6.3KK664 pKa = 8.35WLSLPPHH671 pKa = 6.74LGGAGVTWVTPDD683 pKa = 3.52LTSLALIRR691 pKa = 11.84YY692 pKa = 6.98PLEE695 pKa = 4.5RR696 pKa = 11.84SLLRR700 pKa = 11.84FDD702 pKa = 4.59GVGLFDD708 pKa = 4.37RR709 pKa = 11.84LVSQVDD715 pKa = 3.87LPRR718 pKa = 11.84DD719 pKa = 3.34EE720 pKa = 6.1LYY722 pKa = 10.96AKK724 pKa = 10.06CYY726 pKa = 10.13QYY728 pKa = 11.51LSDD731 pKa = 4.59VVDD734 pKa = 3.45THH736 pKa = 6.18EE737 pKa = 4.79PKK739 pKa = 10.4HH740 pKa = 6.18LYY742 pKa = 10.08RR743 pKa = 11.84DD744 pKa = 3.55ILGSMSKK751 pKa = 8.35PTLDD755 pKa = 2.62WGVPPKK761 pKa = 10.0PWPPLGLVPCGAPPAQPTWARR782 pKa = 11.84GLQFPNWFRR791 pKa = 11.84NIAIDD796 pKa = 3.47VAVRR800 pKa = 11.84TKK802 pKa = 9.01TEE804 pKa = 3.58KK805 pKa = 10.83SVDD808 pKa = 3.66VLFEE812 pKa = 4.71PYY814 pKa = 9.74SASHH818 pKa = 6.01SPIIRR823 pKa = 11.84ARR825 pKa = 11.84HH826 pKa = 5.29GRR828 pKa = 11.84NTWADD833 pKa = 3.3WAKK836 pKa = 10.73GALSLPVRR844 pKa = 11.84CWPGWDD850 pKa = 3.21KK851 pKa = 11.5GYY853 pKa = 10.63LATQYY858 pKa = 10.52RR859 pKa = 11.84ARR861 pKa = 11.84IDD863 pKa = 3.58TFVRR867 pKa = 11.84CYY869 pKa = 10.37LGSSRR874 pKa = 11.84RR875 pKa = 11.84WGSRR879 pKa = 11.84ARR881 pKa = 11.84LASEE885 pKa = 4.01VFFYY889 pKa = 10.79SIQRR893 pKa = 11.84CPDD896 pKa = 2.69SLYY899 pKa = 10.05PYY901 pKa = 10.08RR902 pKa = 11.84IAAA905 pKa = 3.95
Molecular weight: 101.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2974 |
248 |
938 |
594.8 |
64.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.323 ± 0.61 | 1.379 ± 0.41 |
5.077 ± 0.652 | 4.573 ± 0.583 |
2.488 ± 0.363 | 8.171 ± 0.524 |
2.085 ± 0.27 | 3.497 ± 0.475 |
2.757 ± 0.503 | 9.718 ± 0.518 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.816 ± 0.106 | 1.984 ± 0.405 |
8.272 ± 0.654 | 4.035 ± 0.343 |
6.691 ± 0.834 | 7.835 ± 1.151 |
6.153 ± 0.454 | 7.297 ± 0.377 |
2.656 ± 0.396 | 3.194 ± 0.438 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
