
Penicillium digitatum polymycoviruses 1
Taxonomy: Viruses; Riboviria; Polymycoviridae; Polymycovirus; Penicillium digitatum polymycovirus 1
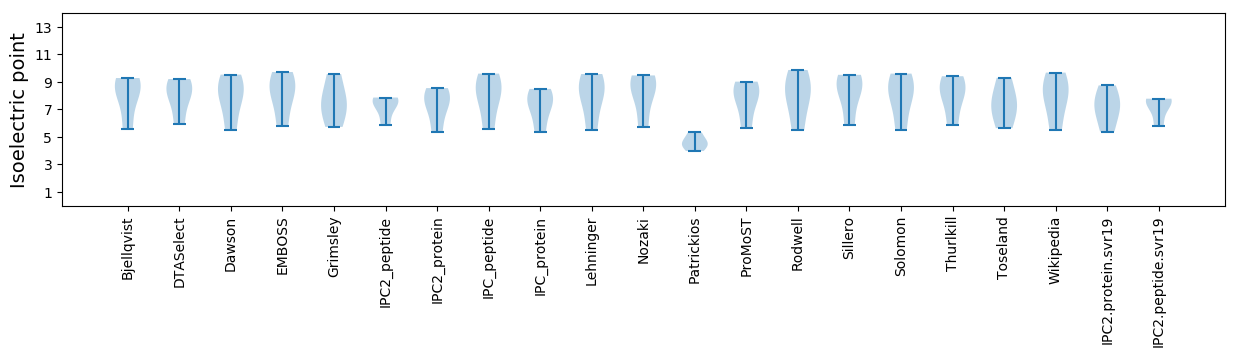
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
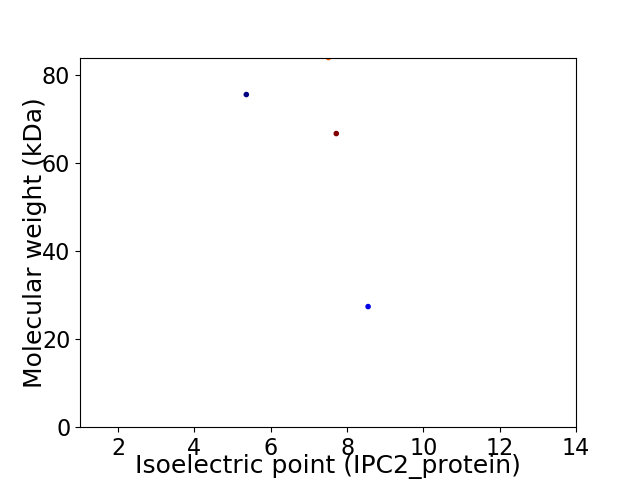
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R4SUG9|A0A2R4SUG9_9VIRU Uncharacterized protein OS=Penicillium digitatum polymycoviruses 1 OX=2164101 PE=4 SV=1
MM1 pKa = 7.68AGNLARR7 pKa = 11.84LRR9 pKa = 11.84SLVLSAHH16 pKa = 6.97ADD18 pKa = 3.67TLLVASTIFHH28 pKa = 6.89CFPDD32 pKa = 4.18PTAHH36 pKa = 6.98QDD38 pKa = 2.76ITRR41 pKa = 11.84PEE43 pKa = 3.7VEE45 pKa = 5.63AYY47 pKa = 9.07INWMASSPYY56 pKa = 8.21PAYY59 pKa = 10.33GRR61 pKa = 11.84VVGHH65 pKa = 6.01VIEE68 pKa = 4.34KK69 pKa = 10.3RR70 pKa = 11.84EE71 pKa = 3.86EE72 pKa = 3.9LVEE75 pKa = 3.84VFMRR79 pKa = 11.84LEE81 pKa = 3.88PEE83 pKa = 4.08RR84 pKa = 11.84YY85 pKa = 9.42EE86 pKa = 5.04DD87 pKa = 4.57VADD90 pKa = 4.3LVNAMSTADD99 pKa = 3.8AAWDD103 pKa = 3.5KK104 pKa = 10.89SARR107 pKa = 11.84ASATNDD113 pKa = 3.13LVRR116 pKa = 11.84NTASAAQLAGHH127 pKa = 5.98SAVEE131 pKa = 4.69FVALEE136 pKa = 4.2SLVSAGYY143 pKa = 10.93DD144 pKa = 3.45NISLATTSIDD154 pKa = 3.7LLRR157 pKa = 11.84KK158 pKa = 8.02TGFIDD163 pKa = 4.63RR164 pKa = 11.84MVDD167 pKa = 3.33DD168 pKa = 5.02GLQLRR173 pKa = 11.84RR174 pKa = 11.84SRR176 pKa = 11.84RR177 pKa = 11.84AVAASWVEE185 pKa = 3.78EE186 pKa = 3.93QTGVRR191 pKa = 11.84RR192 pKa = 11.84FFVSPWCLSKK202 pKa = 10.56QACLALLAHH211 pKa = 6.41YY212 pKa = 10.32VRR214 pKa = 11.84EE215 pKa = 4.15PNRR218 pKa = 11.84ARR220 pKa = 11.84LEE222 pKa = 4.06AVAVRR227 pKa = 11.84RR228 pKa = 11.84GQTALTYY235 pKa = 8.55EE236 pKa = 4.19ARR238 pKa = 11.84AGLQAVRR245 pKa = 11.84AILEE249 pKa = 3.79PAIAILHH256 pKa = 4.64SHH258 pKa = 7.03RR259 pKa = 11.84YY260 pKa = 9.8DD261 pKa = 3.71PLQLAFVDD269 pKa = 3.45RR270 pKa = 11.84HH271 pKa = 4.81GRR273 pKa = 11.84RR274 pKa = 11.84AYY276 pKa = 10.42DD277 pKa = 2.73IATRR281 pKa = 11.84TKK283 pKa = 9.18HH284 pKa = 5.17VLVAFSYY291 pKa = 6.61VHH293 pKa = 6.33CRR295 pKa = 11.84GSEE298 pKa = 3.93DD299 pKa = 3.5TLLEE303 pKa = 4.06LARR306 pKa = 11.84VRR308 pKa = 11.84SALAAPDD315 pKa = 3.12ITACRR320 pKa = 11.84EE321 pKa = 3.74YY322 pKa = 11.47AEE324 pKa = 4.58FRR326 pKa = 11.84DD327 pKa = 3.87AEE329 pKa = 4.4EE330 pKa = 3.98VDD332 pKa = 3.63EE333 pKa = 4.71TGLLVLFARR342 pKa = 11.84TLLATRR348 pKa = 11.84DD349 pKa = 3.54HH350 pKa = 7.19RR351 pKa = 11.84DD352 pKa = 3.6DD353 pKa = 3.56IGFGDD358 pKa = 5.48GYY360 pKa = 11.26NKK362 pKa = 9.84PPCLGLVMRR371 pKa = 11.84ARR373 pKa = 11.84GDD375 pKa = 3.48EE376 pKa = 3.97EE377 pKa = 5.68RR378 pKa = 11.84NTLACVARR386 pKa = 11.84CQEE389 pKa = 3.94TVAYY393 pKa = 9.79LRR395 pKa = 11.84HH396 pKa = 6.27RR397 pKa = 11.84EE398 pKa = 3.82VDD400 pKa = 3.2LGKK403 pKa = 10.25QLLAVHH409 pKa = 6.66WEE411 pKa = 4.06GDD413 pKa = 3.84LNPDD417 pKa = 3.38VVIAAMAVSKK427 pKa = 11.19VDD429 pKa = 3.15IAVDD433 pKa = 3.12IGTASVDD440 pKa = 3.26IPGAAPIGASDD451 pKa = 3.38SSYY454 pKa = 11.54LVLLASAEE462 pKa = 4.05ARR464 pKa = 11.84GLPRR468 pKa = 11.84IIRR471 pKa = 11.84LPYY474 pKa = 8.42PTGLSISDD482 pKa = 3.49RR483 pKa = 11.84VEE485 pKa = 3.77LLYY488 pKa = 10.83EE489 pKa = 4.84FYY491 pKa = 10.05TASGQGAVCYY501 pKa = 10.4VGGGSPFSGKK511 pKa = 9.69PSTQTCADD519 pKa = 3.71ANSVARR525 pKa = 11.84ALAPHH530 pKa = 6.23VLSGALTFGTADD542 pKa = 4.66FILPNLCAAHH552 pKa = 6.36ARR554 pKa = 11.84RR555 pKa = 11.84SEE557 pKa = 3.95VAHH560 pKa = 7.04AIEE563 pKa = 4.72CNGAFEE569 pKa = 4.59YY570 pKa = 10.12TCHH573 pKa = 6.19NCVGFAHH580 pKa = 7.53GLRR583 pKa = 11.84DD584 pKa = 3.65TVEE587 pKa = 4.08LVGMAGARR595 pKa = 11.84LVKK598 pKa = 9.65TRR600 pKa = 11.84SAYY603 pKa = 9.65AHH605 pKa = 6.37NGNFSVEE612 pKa = 4.03LFGAGDD618 pKa = 4.11PDD620 pKa = 5.09FEE622 pKa = 4.9DD623 pKa = 3.71TVQTIEE629 pKa = 3.92TTAFMTAVRR638 pKa = 11.84NADD641 pKa = 3.22WEE643 pKa = 4.71GEE645 pKa = 4.16LVIPEE650 pKa = 4.57RR651 pKa = 11.84GDD653 pKa = 3.33PYY655 pKa = 11.05HH656 pKa = 6.89RR657 pKa = 11.84AFMDD661 pKa = 3.39AMSVATRR668 pKa = 11.84RR669 pKa = 11.84AYY671 pKa = 10.34DD672 pKa = 3.61YY673 pKa = 11.41LSGGEE678 pKa = 4.02VLPVPEE684 pKa = 5.37ADD686 pKa = 3.47LASVARR692 pKa = 11.84SITT695 pKa = 3.32
MM1 pKa = 7.68AGNLARR7 pKa = 11.84LRR9 pKa = 11.84SLVLSAHH16 pKa = 6.97ADD18 pKa = 3.67TLLVASTIFHH28 pKa = 6.89CFPDD32 pKa = 4.18PTAHH36 pKa = 6.98QDD38 pKa = 2.76ITRR41 pKa = 11.84PEE43 pKa = 3.7VEE45 pKa = 5.63AYY47 pKa = 9.07INWMASSPYY56 pKa = 8.21PAYY59 pKa = 10.33GRR61 pKa = 11.84VVGHH65 pKa = 6.01VIEE68 pKa = 4.34KK69 pKa = 10.3RR70 pKa = 11.84EE71 pKa = 3.86EE72 pKa = 3.9LVEE75 pKa = 3.84VFMRR79 pKa = 11.84LEE81 pKa = 3.88PEE83 pKa = 4.08RR84 pKa = 11.84YY85 pKa = 9.42EE86 pKa = 5.04DD87 pKa = 4.57VADD90 pKa = 4.3LVNAMSTADD99 pKa = 3.8AAWDD103 pKa = 3.5KK104 pKa = 10.89SARR107 pKa = 11.84ASATNDD113 pKa = 3.13LVRR116 pKa = 11.84NTASAAQLAGHH127 pKa = 5.98SAVEE131 pKa = 4.69FVALEE136 pKa = 4.2SLVSAGYY143 pKa = 10.93DD144 pKa = 3.45NISLATTSIDD154 pKa = 3.7LLRR157 pKa = 11.84KK158 pKa = 8.02TGFIDD163 pKa = 4.63RR164 pKa = 11.84MVDD167 pKa = 3.33DD168 pKa = 5.02GLQLRR173 pKa = 11.84RR174 pKa = 11.84SRR176 pKa = 11.84RR177 pKa = 11.84AVAASWVEE185 pKa = 3.78EE186 pKa = 3.93QTGVRR191 pKa = 11.84RR192 pKa = 11.84FFVSPWCLSKK202 pKa = 10.56QACLALLAHH211 pKa = 6.41YY212 pKa = 10.32VRR214 pKa = 11.84EE215 pKa = 4.15PNRR218 pKa = 11.84ARR220 pKa = 11.84LEE222 pKa = 4.06AVAVRR227 pKa = 11.84RR228 pKa = 11.84GQTALTYY235 pKa = 8.55EE236 pKa = 4.19ARR238 pKa = 11.84AGLQAVRR245 pKa = 11.84AILEE249 pKa = 3.79PAIAILHH256 pKa = 4.64SHH258 pKa = 7.03RR259 pKa = 11.84YY260 pKa = 9.8DD261 pKa = 3.71PLQLAFVDD269 pKa = 3.45RR270 pKa = 11.84HH271 pKa = 4.81GRR273 pKa = 11.84RR274 pKa = 11.84AYY276 pKa = 10.42DD277 pKa = 2.73IATRR281 pKa = 11.84TKK283 pKa = 9.18HH284 pKa = 5.17VLVAFSYY291 pKa = 6.61VHH293 pKa = 6.33CRR295 pKa = 11.84GSEE298 pKa = 3.93DD299 pKa = 3.5TLLEE303 pKa = 4.06LARR306 pKa = 11.84VRR308 pKa = 11.84SALAAPDD315 pKa = 3.12ITACRR320 pKa = 11.84EE321 pKa = 3.74YY322 pKa = 11.47AEE324 pKa = 4.58FRR326 pKa = 11.84DD327 pKa = 3.87AEE329 pKa = 4.4EE330 pKa = 3.98VDD332 pKa = 3.63EE333 pKa = 4.71TGLLVLFARR342 pKa = 11.84TLLATRR348 pKa = 11.84DD349 pKa = 3.54HH350 pKa = 7.19RR351 pKa = 11.84DD352 pKa = 3.6DD353 pKa = 3.56IGFGDD358 pKa = 5.48GYY360 pKa = 11.26NKK362 pKa = 9.84PPCLGLVMRR371 pKa = 11.84ARR373 pKa = 11.84GDD375 pKa = 3.48EE376 pKa = 3.97EE377 pKa = 5.68RR378 pKa = 11.84NTLACVARR386 pKa = 11.84CQEE389 pKa = 3.94TVAYY393 pKa = 9.79LRR395 pKa = 11.84HH396 pKa = 6.27RR397 pKa = 11.84EE398 pKa = 3.82VDD400 pKa = 3.2LGKK403 pKa = 10.25QLLAVHH409 pKa = 6.66WEE411 pKa = 4.06GDD413 pKa = 3.84LNPDD417 pKa = 3.38VVIAAMAVSKK427 pKa = 11.19VDD429 pKa = 3.15IAVDD433 pKa = 3.12IGTASVDD440 pKa = 3.26IPGAAPIGASDD451 pKa = 3.38SSYY454 pKa = 11.54LVLLASAEE462 pKa = 4.05ARR464 pKa = 11.84GLPRR468 pKa = 11.84IIRR471 pKa = 11.84LPYY474 pKa = 8.42PTGLSISDD482 pKa = 3.49RR483 pKa = 11.84VEE485 pKa = 3.77LLYY488 pKa = 10.83EE489 pKa = 4.84FYY491 pKa = 10.05TASGQGAVCYY501 pKa = 10.4VGGGSPFSGKK511 pKa = 9.69PSTQTCADD519 pKa = 3.71ANSVARR525 pKa = 11.84ALAPHH530 pKa = 6.23VLSGALTFGTADD542 pKa = 4.66FILPNLCAAHH552 pKa = 6.36ARR554 pKa = 11.84RR555 pKa = 11.84SEE557 pKa = 3.95VAHH560 pKa = 7.04AIEE563 pKa = 4.72CNGAFEE569 pKa = 4.59YY570 pKa = 10.12TCHH573 pKa = 6.19NCVGFAHH580 pKa = 7.53GLRR583 pKa = 11.84DD584 pKa = 3.65TVEE587 pKa = 4.08LVGMAGARR595 pKa = 11.84LVKK598 pKa = 9.65TRR600 pKa = 11.84SAYY603 pKa = 9.65AHH605 pKa = 6.37NGNFSVEE612 pKa = 4.03LFGAGDD618 pKa = 4.11PDD620 pKa = 5.09FEE622 pKa = 4.9DD623 pKa = 3.71TVQTIEE629 pKa = 3.92TTAFMTAVRR638 pKa = 11.84NADD641 pKa = 3.22WEE643 pKa = 4.71GEE645 pKa = 4.16LVIPEE650 pKa = 4.57RR651 pKa = 11.84GDD653 pKa = 3.33PYY655 pKa = 11.05HH656 pKa = 6.89RR657 pKa = 11.84AFMDD661 pKa = 3.39AMSVATRR668 pKa = 11.84RR669 pKa = 11.84AYY671 pKa = 10.34DD672 pKa = 3.61YY673 pKa = 11.41LSGGEE678 pKa = 4.02VLPVPEE684 pKa = 5.37ADD686 pKa = 3.47LASVARR692 pKa = 11.84SITT695 pKa = 3.32
Molecular weight: 75.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R4SUG9|A0A2R4SUG9_9VIRU Uncharacterized protein OS=Penicillium digitatum polymycoviruses 1 OX=2164101 PE=4 SV=1
MM1 pKa = 7.7SSPTAPRR8 pKa = 11.84LVSVEE13 pKa = 3.99KK14 pKa = 10.88AKK16 pKa = 11.05LIADD20 pKa = 4.64LDD22 pKa = 3.85QSVVDD27 pKa = 4.29VILRR31 pKa = 11.84LSSLGVKK38 pKa = 9.77PDD40 pKa = 4.83GIIDD44 pKa = 3.58YY45 pKa = 9.78CNRR48 pKa = 11.84VALDD52 pKa = 3.8EE53 pKa = 4.38PAPAVVAGGAKK64 pKa = 9.96PLVIQAYY71 pKa = 10.23SFLSDD76 pKa = 4.39KK77 pKa = 10.79SICVDD82 pKa = 3.09KK83 pKa = 11.22FGMEE87 pKa = 4.3LSAAAAALEE96 pKa = 4.1TLKK99 pKa = 10.48RR100 pKa = 11.84DD101 pKa = 3.56PEE103 pKa = 3.88EE104 pKa = 3.95GRR106 pKa = 11.84RR107 pKa = 11.84IVHH110 pKa = 6.47KK111 pKa = 10.74AVADD115 pKa = 3.86HH116 pKa = 4.94QAKK119 pKa = 10.21RR120 pKa = 11.84GSPKK124 pKa = 10.29PIVVNLAGLPSSRR137 pKa = 11.84GGPKK141 pKa = 9.69TGGDD145 pKa = 3.27GGKK148 pKa = 10.26AEE150 pKa = 4.62LSEE153 pKa = 4.38LMRR156 pKa = 11.84RR157 pKa = 11.84NQALAGAYY165 pKa = 9.69AFVAEE170 pKa = 4.43EE171 pKa = 4.39HH172 pKa = 6.66GPPGPEE178 pKa = 3.46RR179 pKa = 11.84FRR181 pKa = 11.84VLLGGNLAVVGQNKK195 pKa = 9.51ACAVKK200 pKa = 10.18AAALVRR206 pKa = 11.84ILGRR210 pKa = 11.84RR211 pKa = 11.84HH212 pKa = 3.8QTLVKK217 pKa = 9.72YY218 pKa = 10.23VRR220 pKa = 11.84GYY222 pKa = 10.53IPGKK226 pKa = 10.5ALLPSDD232 pKa = 5.25KK233 pKa = 10.2IPPLIYY239 pKa = 10.46DD240 pKa = 4.51GIIAPDD246 pKa = 3.77EE247 pKa = 4.5KK248 pKa = 10.88PPSDD252 pKa = 3.6EE253 pKa = 4.89SKK255 pKa = 10.72GQSTVGG261 pKa = 3.15
MM1 pKa = 7.7SSPTAPRR8 pKa = 11.84LVSVEE13 pKa = 3.99KK14 pKa = 10.88AKK16 pKa = 11.05LIADD20 pKa = 4.64LDD22 pKa = 3.85QSVVDD27 pKa = 4.29VILRR31 pKa = 11.84LSSLGVKK38 pKa = 9.77PDD40 pKa = 4.83GIIDD44 pKa = 3.58YY45 pKa = 9.78CNRR48 pKa = 11.84VALDD52 pKa = 3.8EE53 pKa = 4.38PAPAVVAGGAKK64 pKa = 9.96PLVIQAYY71 pKa = 10.23SFLSDD76 pKa = 4.39KK77 pKa = 10.79SICVDD82 pKa = 3.09KK83 pKa = 11.22FGMEE87 pKa = 4.3LSAAAAALEE96 pKa = 4.1TLKK99 pKa = 10.48RR100 pKa = 11.84DD101 pKa = 3.56PEE103 pKa = 3.88EE104 pKa = 3.95GRR106 pKa = 11.84RR107 pKa = 11.84IVHH110 pKa = 6.47KK111 pKa = 10.74AVADD115 pKa = 3.86HH116 pKa = 4.94QAKK119 pKa = 10.21RR120 pKa = 11.84GSPKK124 pKa = 10.29PIVVNLAGLPSSRR137 pKa = 11.84GGPKK141 pKa = 9.69TGGDD145 pKa = 3.27GGKK148 pKa = 10.26AEE150 pKa = 4.62LSEE153 pKa = 4.38LMRR156 pKa = 11.84RR157 pKa = 11.84NQALAGAYY165 pKa = 9.69AFVAEE170 pKa = 4.43EE171 pKa = 4.39HH172 pKa = 6.66GPPGPEE178 pKa = 3.46RR179 pKa = 11.84FRR181 pKa = 11.84VLLGGNLAVVGQNKK195 pKa = 9.51ACAVKK200 pKa = 10.18AAALVRR206 pKa = 11.84ILGRR210 pKa = 11.84RR211 pKa = 11.84HH212 pKa = 3.8QTLVKK217 pKa = 9.72YY218 pKa = 10.23VRR220 pKa = 11.84GYY222 pKa = 10.53IPGKK226 pKa = 10.5ALLPSDD232 pKa = 5.25KK233 pKa = 10.2IPPLIYY239 pKa = 10.46DD240 pKa = 4.51GIIAPDD246 pKa = 3.77EE247 pKa = 4.5KK248 pKa = 10.88PPSDD252 pKa = 3.6EE253 pKa = 4.89SKK255 pKa = 10.72GQSTVGG261 pKa = 3.15
Molecular weight: 27.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2328 |
261 |
760 |
582.0 |
63.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.868 ± 1.516 | 1.375 ± 0.247 |
6.271 ± 0.085 | 5.498 ± 0.361 |
3.393 ± 0.457 | 8.333 ± 0.849 |
2.706 ± 0.325 | 3.179 ± 0.494 |
3.351 ± 0.823 | 9.622 ± 0.436 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.105 ± 0.329 | 2.448 ± 0.277 |
5.412 ± 0.869 | 2.448 ± 0.241 |
8.247 ± 0.345 | 6.787 ± 0.286 |
4.94 ± 0.685 | 9.149 ± 0.328 |
0.816 ± 0.135 | 3.05 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
