
Candidatus Magnetobacterium bavaricum
Taxonomy: cellular organisms; Bacteria; Nitrospirae; Nitrospira; Nitrospirales; Nitrospiraceae; Candidatus Magnetobacterium
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
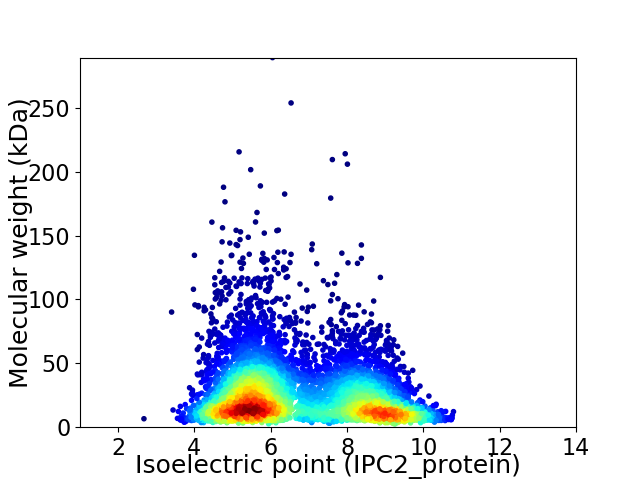
Virtual 2D-PAGE plot for 6447 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F3GVL4|A0A0F3GVL4_9BACT Uncharacterized protein OS=Candidatus Magnetobacterium bavaricum OX=29290 GN=MBAV_001914 PE=4 SV=1
MM1 pKa = 7.74HH2 pKa = 7.92DD3 pKa = 4.4IISLSLQDD11 pKa = 3.95YY12 pKa = 9.4PDD14 pKa = 3.86VVGDD18 pKa = 4.83DD19 pKa = 3.82YY20 pKa = 10.7WTSTDD25 pKa = 2.99SDD27 pKa = 3.59IYY29 pKa = 9.8DD30 pKa = 3.54TFVIGMSSGFVSSSSRR46 pKa = 11.84TYY48 pKa = 10.59PNSVWPVRR56 pKa = 11.84SGQQ59 pKa = 3.25
MM1 pKa = 7.74HH2 pKa = 7.92DD3 pKa = 4.4IISLSLQDD11 pKa = 3.95YY12 pKa = 9.4PDD14 pKa = 3.86VVGDD18 pKa = 4.83DD19 pKa = 3.82YY20 pKa = 10.7WTSTDD25 pKa = 2.99SDD27 pKa = 3.59IYY29 pKa = 9.8DD30 pKa = 3.54TFVIGMSSGFVSSSSRR46 pKa = 11.84TYY48 pKa = 10.59PNSVWPVRR56 pKa = 11.84SGQQ59 pKa = 3.25
Molecular weight: 6.58 kDa
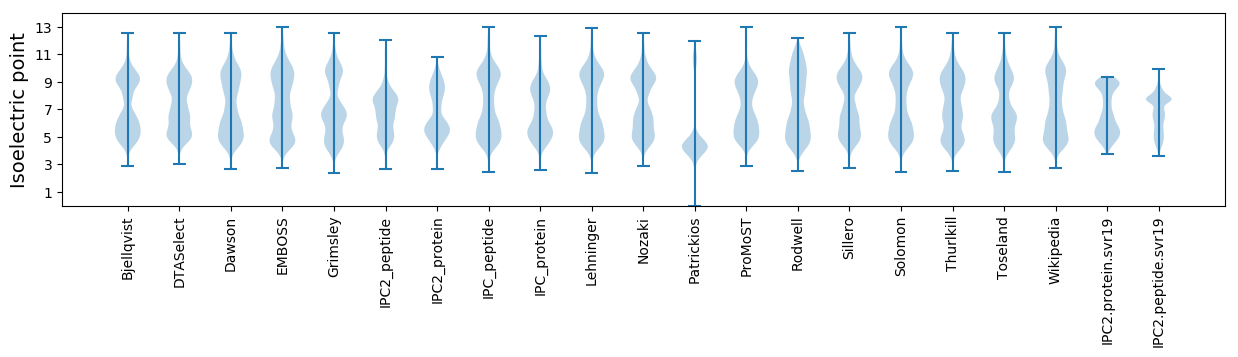
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F3GU45|A0A0F3GU45_9BACT Uncharacterized protein OS=Candidatus Magnetobacterium bavaricum OX=29290 GN=MBAV_003569 PE=4 SV=1
MM1 pKa = 7.73EE2 pKa = 5.5PFSYY6 pKa = 10.3QPRR9 pKa = 11.84HH10 pKa = 6.21CSHH13 pKa = 6.46CRR15 pKa = 11.84CRR17 pKa = 11.84IGGDD21 pKa = 3.13KK22 pKa = 10.77RR23 pKa = 11.84QGSQSARR30 pKa = 11.84TQCAARR36 pKa = 11.84VKK38 pKa = 9.8PKK40 pKa = 9.98PSKK43 pKa = 8.72PQQRR47 pKa = 11.84PTQDD51 pKa = 2.54RR52 pKa = 11.84QGQVVRR58 pKa = 11.84GHH60 pKa = 5.88VFPSKK65 pKa = 10.85APAFAHH71 pKa = 7.14DD72 pKa = 4.24NEE74 pKa = 5.14CGHH77 pKa = 6.71GGHH80 pKa = 6.59CRR82 pKa = 11.84VYY84 pKa = 9.41MHH86 pKa = 7.39DD87 pKa = 3.48RR88 pKa = 11.84TSGKK92 pKa = 9.51VQGTEE97 pKa = 3.75LCHH100 pKa = 5.99YY101 pKa = 8.15PAAAPHH107 pKa = 6.59PMAQGRR113 pKa = 11.84IHH115 pKa = 6.91KK116 pKa = 8.66RR117 pKa = 11.84QPQRR121 pKa = 11.84RR122 pKa = 11.84KK123 pKa = 7.71QQQGRR128 pKa = 11.84EE129 pKa = 4.01LHH131 pKa = 6.3PLGKK135 pKa = 8.57RR136 pKa = 11.84TRR138 pKa = 11.84YY139 pKa = 7.97QRR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84YY144 pKa = 9.19DD145 pKa = 3.29RR146 pKa = 11.84KK147 pKa = 10.1HH148 pKa = 5.92PLIHH152 pKa = 7.63HH153 pKa = 6.42EE154 pKa = 4.05EE155 pKa = 4.32QVWNRR160 pKa = 11.84LCVIRR165 pKa = 11.84RR166 pKa = 11.84LLSDD170 pKa = 3.9RR171 pKa = 11.84VQPKK175 pKa = 8.77PRR177 pKa = 11.84KK178 pKa = 9.51RR179 pKa = 11.84PNHH182 pKa = 5.91PANVRR187 pKa = 11.84PKK189 pKa = 9.94GQTITPQHH197 pKa = 6.97PYY199 pKa = 8.43HH200 pKa = 6.84TDD202 pKa = 2.91HH203 pKa = 6.51AQHH206 pKa = 6.16YY207 pKa = 8.48KK208 pKa = 10.42AVHH211 pKa = 5.87NSAKK215 pKa = 10.43DD216 pKa = 3.51VLFSDD221 pKa = 4.28HH222 pKa = 6.92PAVEE226 pKa = 4.62KK227 pKa = 10.68SQSGGHH233 pKa = 5.16QHH235 pKa = 5.98HH236 pKa = 6.3QCGTYY241 pKa = 9.65QEE243 pKa = 4.69PGGIAGVNLEE253 pKa = 4.13HH254 pKa = 7.31LLWCLRR260 pKa = 11.84GQGRR264 pKa = 11.84RR265 pKa = 11.84AA266 pKa = 3.14
MM1 pKa = 7.73EE2 pKa = 5.5PFSYY6 pKa = 10.3QPRR9 pKa = 11.84HH10 pKa = 6.21CSHH13 pKa = 6.46CRR15 pKa = 11.84CRR17 pKa = 11.84IGGDD21 pKa = 3.13KK22 pKa = 10.77RR23 pKa = 11.84QGSQSARR30 pKa = 11.84TQCAARR36 pKa = 11.84VKK38 pKa = 9.8PKK40 pKa = 9.98PSKK43 pKa = 8.72PQQRR47 pKa = 11.84PTQDD51 pKa = 2.54RR52 pKa = 11.84QGQVVRR58 pKa = 11.84GHH60 pKa = 5.88VFPSKK65 pKa = 10.85APAFAHH71 pKa = 7.14DD72 pKa = 4.24NEE74 pKa = 5.14CGHH77 pKa = 6.71GGHH80 pKa = 6.59CRR82 pKa = 11.84VYY84 pKa = 9.41MHH86 pKa = 7.39DD87 pKa = 3.48RR88 pKa = 11.84TSGKK92 pKa = 9.51VQGTEE97 pKa = 3.75LCHH100 pKa = 5.99YY101 pKa = 8.15PAAAPHH107 pKa = 6.59PMAQGRR113 pKa = 11.84IHH115 pKa = 6.91KK116 pKa = 8.66RR117 pKa = 11.84QPQRR121 pKa = 11.84RR122 pKa = 11.84KK123 pKa = 7.71QQQGRR128 pKa = 11.84EE129 pKa = 4.01LHH131 pKa = 6.3PLGKK135 pKa = 8.57RR136 pKa = 11.84TRR138 pKa = 11.84YY139 pKa = 7.97QRR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84YY144 pKa = 9.19DD145 pKa = 3.29RR146 pKa = 11.84KK147 pKa = 10.1HH148 pKa = 5.92PLIHH152 pKa = 7.63HH153 pKa = 6.42EE154 pKa = 4.05EE155 pKa = 4.32QVWNRR160 pKa = 11.84LCVIRR165 pKa = 11.84RR166 pKa = 11.84LLSDD170 pKa = 3.9RR171 pKa = 11.84VQPKK175 pKa = 8.77PRR177 pKa = 11.84KK178 pKa = 9.51RR179 pKa = 11.84PNHH182 pKa = 5.91PANVRR187 pKa = 11.84PKK189 pKa = 9.94GQTITPQHH197 pKa = 6.97PYY199 pKa = 8.43HH200 pKa = 6.84TDD202 pKa = 2.91HH203 pKa = 6.51AQHH206 pKa = 6.16YY207 pKa = 8.48KK208 pKa = 10.42AVHH211 pKa = 5.87NSAKK215 pKa = 10.43DD216 pKa = 3.51VLFSDD221 pKa = 4.28HH222 pKa = 6.92PAVEE226 pKa = 4.62KK227 pKa = 10.68SQSGGHH233 pKa = 5.16QHH235 pKa = 5.98HH236 pKa = 6.3QCGTYY241 pKa = 9.65QEE243 pKa = 4.69PGGIAGVNLEE253 pKa = 4.13HH254 pKa = 7.31LLWCLRR260 pKa = 11.84GQGRR264 pKa = 11.84RR265 pKa = 11.84AA266 pKa = 3.14
Molecular weight: 30.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1584456 |
22 |
2585 |
245.8 |
27.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.179 ± 0.036 | 1.302 ± 0.015 |
5.931 ± 0.031 | 5.714 ± 0.043 |
4.106 ± 0.028 | 7.058 ± 0.041 |
1.924 ± 0.017 | 7.537 ± 0.034 |
6.315 ± 0.036 | 9.37 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.725 ± 0.015 | 4.446 ± 0.029 |
3.829 ± 0.024 | 3.084 ± 0.017 |
5.009 ± 0.036 | 6.508 ± 0.037 |
6.038 ± 0.05 | 7.206 ± 0.03 |
1.101 ± 0.021 | 3.616 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
