
Grapevine leafroll-associated virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Ampelovirus
Average proteome isoelectric point is 7.11
Get precalculated fractions of proteins
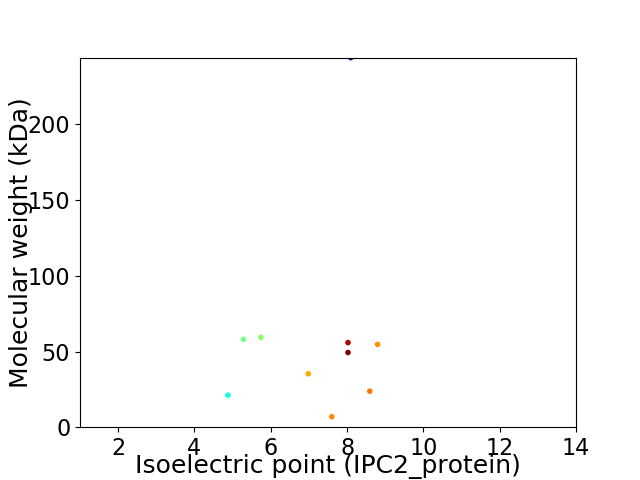
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G9L8F5|G9L8F5_9CLOS p24 OS=Grapevine leafroll-associated virus 1 OX=47985 PE=4 SV=1
MM1 pKa = 7.42EE2 pKa = 5.49FAPVCYY8 pKa = 9.69ICTKK12 pKa = 10.65NEE14 pKa = 3.69EE15 pKa = 4.25GSDD18 pKa = 3.52KK19 pKa = 10.96KK20 pKa = 10.59LTLLSRR26 pKa = 11.84EE27 pKa = 4.17DD28 pKa = 4.95LIDD31 pKa = 4.13FNSDD35 pKa = 3.05EE36 pKa = 4.46NQDD39 pKa = 3.69EE40 pKa = 4.36LSLEE44 pKa = 3.94KK45 pKa = 10.79KK46 pKa = 10.23PFVKK50 pKa = 10.26SYY52 pKa = 11.07EE53 pKa = 3.93NYY55 pKa = 10.09LRR57 pKa = 11.84DD58 pKa = 3.15WAEE61 pKa = 3.66RR62 pKa = 11.84PFYY65 pKa = 10.33FSNAMSVMDD74 pKa = 4.73LAMHH78 pKa = 6.97TGNASIDD85 pKa = 3.76SDD87 pKa = 3.9SGLSKK92 pKa = 10.64SGEE95 pKa = 3.92KK96 pKa = 10.04RR97 pKa = 11.84RR98 pKa = 11.84TAVEE102 pKa = 4.01VQSPWKK108 pKa = 9.39TGIPFDD114 pKa = 4.26SVDD117 pKa = 3.46NGVLVAVGYY126 pKa = 10.34EE127 pKa = 4.03GLNADD132 pKa = 3.78KK133 pKa = 10.92KK134 pKa = 9.73KK135 pKa = 10.64ARR137 pKa = 11.84SVALFMQNEE146 pKa = 4.44GSYY149 pKa = 11.1IMLIGGEE156 pKa = 4.69VYY158 pKa = 10.74ASSSYY163 pKa = 10.54IKK165 pKa = 10.55EE166 pKa = 4.0CMRR169 pKa = 11.84CQRR172 pKa = 11.84LASTMSGDD180 pKa = 3.76VFVDD184 pKa = 3.51ALRR187 pKa = 11.84LVV189 pKa = 3.58
MM1 pKa = 7.42EE2 pKa = 5.49FAPVCYY8 pKa = 9.69ICTKK12 pKa = 10.65NEE14 pKa = 3.69EE15 pKa = 4.25GSDD18 pKa = 3.52KK19 pKa = 10.96KK20 pKa = 10.59LTLLSRR26 pKa = 11.84EE27 pKa = 4.17DD28 pKa = 4.95LIDD31 pKa = 4.13FNSDD35 pKa = 3.05EE36 pKa = 4.46NQDD39 pKa = 3.69EE40 pKa = 4.36LSLEE44 pKa = 3.94KK45 pKa = 10.79KK46 pKa = 10.23PFVKK50 pKa = 10.26SYY52 pKa = 11.07EE53 pKa = 3.93NYY55 pKa = 10.09LRR57 pKa = 11.84DD58 pKa = 3.15WAEE61 pKa = 3.66RR62 pKa = 11.84PFYY65 pKa = 10.33FSNAMSVMDD74 pKa = 4.73LAMHH78 pKa = 6.97TGNASIDD85 pKa = 3.76SDD87 pKa = 3.9SGLSKK92 pKa = 10.64SGEE95 pKa = 3.92KK96 pKa = 10.04RR97 pKa = 11.84RR98 pKa = 11.84TAVEE102 pKa = 4.01VQSPWKK108 pKa = 9.39TGIPFDD114 pKa = 4.26SVDD117 pKa = 3.46NGVLVAVGYY126 pKa = 10.34EE127 pKa = 4.03GLNADD132 pKa = 3.78KK133 pKa = 10.92KK134 pKa = 9.73KK135 pKa = 10.64ARR137 pKa = 11.84SVALFMQNEE146 pKa = 4.44GSYY149 pKa = 11.1IMLIGGEE156 pKa = 4.69VYY158 pKa = 10.74ASSSYY163 pKa = 10.54IKK165 pKa = 10.55EE166 pKa = 4.0CMRR169 pKa = 11.84CQRR172 pKa = 11.84LASTMSGDD180 pKa = 3.76VFVDD184 pKa = 3.51ALRR187 pKa = 11.84LVV189 pKa = 3.58
Molecular weight: 21.15 kDa
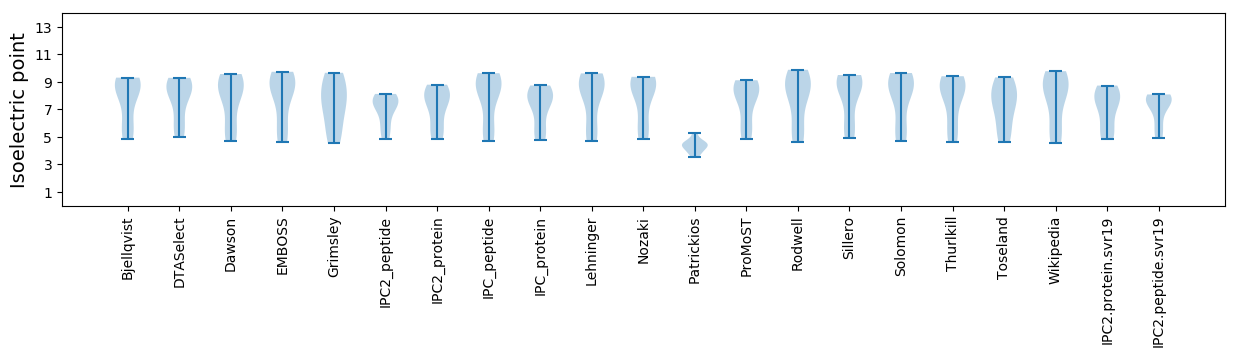
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G9L8F1|G9L8F1_9CLOS Coat protein OS=Grapevine leafroll-associated virus 1 OX=47985 GN=CP PE=4 SV=1
MM1 pKa = 7.4NPLVWLTGATDD12 pKa = 3.55YY13 pKa = 11.1MSVLRR18 pKa = 11.84PLYY21 pKa = 10.36EE22 pKa = 4.16DD23 pKa = 4.65FKK25 pKa = 11.28AGGGRR30 pKa = 11.84RR31 pKa = 11.84VVNYY35 pKa = 10.07IEE37 pKa = 4.63VIDD40 pKa = 5.1RR41 pKa = 11.84GDD43 pKa = 3.3KK44 pKa = 9.24PHH46 pKa = 7.63IIRR49 pKa = 11.84GLSPSDD55 pKa = 3.43EE56 pKa = 4.18NFEE59 pKa = 4.64LIAWAVAVGAFEE71 pKa = 3.9NSIYY75 pKa = 11.07AFDD78 pKa = 4.96TILSTMRR85 pKa = 11.84NWKK88 pKa = 10.23AYY90 pKa = 6.83ATPGSLQLDD99 pKa = 3.79TPITKK104 pKa = 10.16KK105 pKa = 10.44KK106 pKa = 10.0NKK108 pKa = 9.73HH109 pKa = 4.32FTLTAEE115 pKa = 3.81QQAAVRR121 pKa = 11.84TSPNSTNEE129 pKa = 3.32AFAQLFLNLGVFLKK143 pKa = 10.38RR144 pKa = 11.84VPKK147 pKa = 10.82DD148 pKa = 3.23EE149 pKa = 5.67DD150 pKa = 3.07ISGRR154 pKa = 11.84GNIPLVRR161 pKa = 11.84VSQAEE166 pKa = 3.99STDD169 pKa = 3.72RR170 pKa = 11.84IEE172 pKa = 3.99QGRR175 pKa = 11.84YY176 pKa = 9.45SPLLAKK182 pKa = 9.8FVRR185 pKa = 11.84KK186 pKa = 7.94YY187 pKa = 7.68THH189 pKa = 6.49VVDD192 pKa = 5.59AYY194 pKa = 10.64DD195 pKa = 4.36SVVTRR200 pKa = 11.84QKK202 pKa = 10.61IEE204 pKa = 3.73VCSYY208 pKa = 9.47YY209 pKa = 10.03IPEE212 pKa = 5.14LIGLLFRR219 pKa = 11.84ARR221 pKa = 11.84DD222 pKa = 3.26GAQAYY227 pKa = 6.65KK228 pKa = 10.05TVSLYY233 pKa = 10.7LASFLQSKK241 pKa = 10.79DD242 pKa = 3.05PGKK245 pKa = 10.5YY246 pKa = 9.92DD247 pKa = 3.28PEE249 pKa = 4.12TRR251 pKa = 11.84VIWFARR257 pKa = 11.84TLTMARR263 pKa = 11.84EE264 pKa = 3.99LLEE267 pKa = 3.68IYY269 pKa = 10.71YY270 pKa = 9.95NIRR273 pKa = 11.84VEE275 pKa = 4.0VSEE278 pKa = 4.8RR279 pKa = 11.84EE280 pKa = 3.74IIRR283 pKa = 11.84NFRR286 pKa = 11.84IQPGDD291 pKa = 3.62EE292 pKa = 3.91FHH294 pKa = 6.72QLSTMLLGLVADD306 pKa = 5.05TSAKK310 pKa = 9.23LTGITDD316 pKa = 3.11VDD318 pKa = 3.94VVLRR322 pKa = 11.84RR323 pKa = 11.84VLGLCLKK330 pKa = 11.08AMVDD334 pKa = 4.07EE335 pKa = 4.73NQHH338 pKa = 5.67ISNINYY344 pKa = 8.45TGALYY349 pKa = 11.22LMLVYY354 pKa = 9.48FSIVGTNYY362 pKa = 9.72LRR364 pKa = 11.84KK365 pKa = 9.55KK366 pKa = 9.11DD367 pKa = 3.99TPNEE371 pKa = 3.73MVFRR375 pKa = 11.84INGVVRR381 pKa = 11.84KK382 pKa = 10.48VNFTHH387 pKa = 7.75LKK389 pKa = 9.77RR390 pKa = 11.84LVAEE394 pKa = 4.33ASVDD398 pKa = 3.01GRR400 pKa = 11.84NYY402 pKa = 9.37PRR404 pKa = 11.84EE405 pKa = 3.91ISSVFAGITLQLRR418 pKa = 11.84QLGNIDD424 pKa = 3.57TVVWPDD430 pKa = 3.43VVLSNPSLGFDD441 pKa = 3.6TVLHH445 pKa = 6.18SGYY448 pKa = 10.19ISLNPAMYY456 pKa = 10.5KK457 pKa = 10.28DD458 pKa = 3.25IEE460 pKa = 4.37IIKK463 pKa = 10.25RR464 pKa = 11.84KK465 pKa = 9.18IRR467 pKa = 11.84NSTHH471 pKa = 4.88WVGGYY476 pKa = 10.32KK477 pKa = 9.83YY478 pKa = 10.72GKK480 pKa = 10.09RR481 pKa = 11.84FF482 pKa = 3.22
MM1 pKa = 7.4NPLVWLTGATDD12 pKa = 3.55YY13 pKa = 11.1MSVLRR18 pKa = 11.84PLYY21 pKa = 10.36EE22 pKa = 4.16DD23 pKa = 4.65FKK25 pKa = 11.28AGGGRR30 pKa = 11.84RR31 pKa = 11.84VVNYY35 pKa = 10.07IEE37 pKa = 4.63VIDD40 pKa = 5.1RR41 pKa = 11.84GDD43 pKa = 3.3KK44 pKa = 9.24PHH46 pKa = 7.63IIRR49 pKa = 11.84GLSPSDD55 pKa = 3.43EE56 pKa = 4.18NFEE59 pKa = 4.64LIAWAVAVGAFEE71 pKa = 3.9NSIYY75 pKa = 11.07AFDD78 pKa = 4.96TILSTMRR85 pKa = 11.84NWKK88 pKa = 10.23AYY90 pKa = 6.83ATPGSLQLDD99 pKa = 3.79TPITKK104 pKa = 10.16KK105 pKa = 10.44KK106 pKa = 10.0NKK108 pKa = 9.73HH109 pKa = 4.32FTLTAEE115 pKa = 3.81QQAAVRR121 pKa = 11.84TSPNSTNEE129 pKa = 3.32AFAQLFLNLGVFLKK143 pKa = 10.38RR144 pKa = 11.84VPKK147 pKa = 10.82DD148 pKa = 3.23EE149 pKa = 5.67DD150 pKa = 3.07ISGRR154 pKa = 11.84GNIPLVRR161 pKa = 11.84VSQAEE166 pKa = 3.99STDD169 pKa = 3.72RR170 pKa = 11.84IEE172 pKa = 3.99QGRR175 pKa = 11.84YY176 pKa = 9.45SPLLAKK182 pKa = 9.8FVRR185 pKa = 11.84KK186 pKa = 7.94YY187 pKa = 7.68THH189 pKa = 6.49VVDD192 pKa = 5.59AYY194 pKa = 10.64DD195 pKa = 4.36SVVTRR200 pKa = 11.84QKK202 pKa = 10.61IEE204 pKa = 3.73VCSYY208 pKa = 9.47YY209 pKa = 10.03IPEE212 pKa = 5.14LIGLLFRR219 pKa = 11.84ARR221 pKa = 11.84DD222 pKa = 3.26GAQAYY227 pKa = 6.65KK228 pKa = 10.05TVSLYY233 pKa = 10.7LASFLQSKK241 pKa = 10.79DD242 pKa = 3.05PGKK245 pKa = 10.5YY246 pKa = 9.92DD247 pKa = 3.28PEE249 pKa = 4.12TRR251 pKa = 11.84VIWFARR257 pKa = 11.84TLTMARR263 pKa = 11.84EE264 pKa = 3.99LLEE267 pKa = 3.68IYY269 pKa = 10.71YY270 pKa = 9.95NIRR273 pKa = 11.84VEE275 pKa = 4.0VSEE278 pKa = 4.8RR279 pKa = 11.84EE280 pKa = 3.74IIRR283 pKa = 11.84NFRR286 pKa = 11.84IQPGDD291 pKa = 3.62EE292 pKa = 3.91FHH294 pKa = 6.72QLSTMLLGLVADD306 pKa = 5.05TSAKK310 pKa = 9.23LTGITDD316 pKa = 3.11VDD318 pKa = 3.94VVLRR322 pKa = 11.84RR323 pKa = 11.84VLGLCLKK330 pKa = 11.08AMVDD334 pKa = 4.07EE335 pKa = 4.73NQHH338 pKa = 5.67ISNINYY344 pKa = 8.45TGALYY349 pKa = 11.22LMLVYY354 pKa = 9.48FSIVGTNYY362 pKa = 9.72LRR364 pKa = 11.84KK365 pKa = 9.55KK366 pKa = 9.11DD367 pKa = 3.99TPNEE371 pKa = 3.73MVFRR375 pKa = 11.84INGVVRR381 pKa = 11.84KK382 pKa = 10.48VNFTHH387 pKa = 7.75LKK389 pKa = 9.77RR390 pKa = 11.84LVAEE394 pKa = 4.33ASVDD398 pKa = 3.01GRR400 pKa = 11.84NYY402 pKa = 9.37PRR404 pKa = 11.84EE405 pKa = 3.91ISSVFAGITLQLRR418 pKa = 11.84QLGNIDD424 pKa = 3.57TVVWPDD430 pKa = 3.43VVLSNPSLGFDD441 pKa = 3.6TVLHH445 pKa = 6.18SGYY448 pKa = 10.19ISLNPAMYY456 pKa = 10.5KK457 pKa = 10.28DD458 pKa = 3.25IEE460 pKa = 4.37IIKK463 pKa = 10.25RR464 pKa = 11.84KK465 pKa = 9.18IRR467 pKa = 11.84NSTHH471 pKa = 4.88WVGGYY476 pKa = 10.32KK477 pKa = 9.83YY478 pKa = 10.72GKK480 pKa = 10.09RR481 pKa = 11.84FF482 pKa = 3.22
Molecular weight: 54.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5466 |
59 |
2204 |
546.6 |
60.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.245 ± 0.415 | 1.5 ± 0.317 |
5.671 ± 0.268 | 6.092 ± 0.255 |
4.409 ± 0.364 | 6.604 ± 0.357 |
1.518 ± 0.18 | 5.123 ± 0.453 |
6.495 ± 0.359 | 9.239 ± 0.431 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.141 ± 0.128 | 4.135 ± 0.414 |
3.531 ± 0.319 | 2.47 ± 0.176 |
6.147 ± 0.284 | 8.361 ± 0.27 |
5.507 ± 0.309 | 9.276 ± 0.305 |
0.732 ± 0.095 | 3.805 ± 0.323 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
