
Rattus norvegicus papillomavirus 1 (isolate Rat/Germany/Schulz/2009) (RnPV-1)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Pipapillomavirus; Pipapillomavirus 2
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
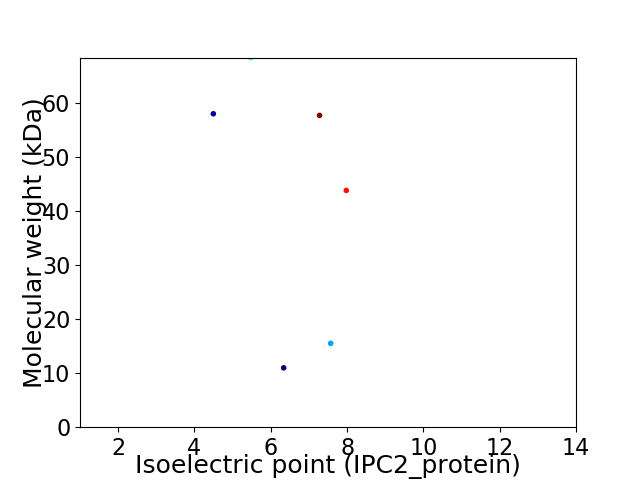
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
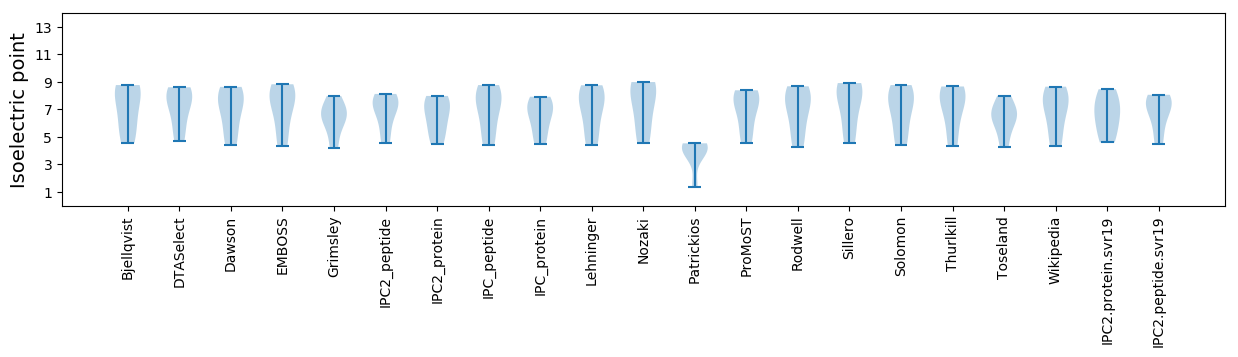
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C8AY94|C8AY94_RNPV1 Major capsid protein L1 OS=Rattus norvegicus papillomavirus 1 (isolate Rat/Germany/Schulz/2009) OX=664730 GN=L1 PE=3 SV=1
MM1 pKa = 7.25VSATRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VKK10 pKa = 10.34RR11 pKa = 11.84DD12 pKa = 3.07SASNLYY18 pKa = 9.65RR19 pKa = 11.84QCQVTGNCPPDD30 pKa = 3.41VANKK34 pKa = 10.19IEE36 pKa = 4.27GNTLADD42 pKa = 3.59KK43 pKa = 10.53LLKK46 pKa = 10.12IFSSIIYY53 pKa = 10.17LGGLGIGTGRR63 pKa = 11.84GTGEE67 pKa = 4.0TYY69 pKa = 10.56GYY71 pKa = 11.24GPINPGGGRR80 pKa = 11.84ITGTGTVMRR89 pKa = 11.84PGVVVEE95 pKa = 4.83PIGPAEE101 pKa = 4.09IVPVDD106 pKa = 3.64SVGPGEE112 pKa = 4.24SSIVPLLEE120 pKa = 4.05ATPDD124 pKa = 3.45VPINGGPEE132 pKa = 3.87VPPAGPDD139 pKa = 3.14TSTVDD144 pKa = 3.23VTSNIDD150 pKa = 3.32PVSDD154 pKa = 3.75VNVSGGTTITNTDD167 pKa = 3.28SAVIDD172 pKa = 3.9VQPAPSGPRR181 pKa = 11.84RR182 pKa = 11.84VTVTRR187 pKa = 11.84SEE189 pKa = 4.53HH190 pKa = 6.01NNPAYY195 pKa = 10.68VSVSAPPQGLGEE207 pKa = 4.39GGGFLLAGSEE217 pKa = 4.26SSGLIASTHH226 pKa = 5.62EE227 pKa = 3.93LDD229 pKa = 3.42AAVIIGGRR237 pKa = 11.84PPPEE241 pKa = 4.2TIFEE245 pKa = 4.98DD246 pKa = 3.55IEE248 pKa = 5.74LDD250 pKa = 3.65TFSATGGRR258 pKa = 11.84SEE260 pKa = 4.46FDD262 pKa = 2.76IEE264 pKa = 4.33EE265 pKa = 4.99PYY267 pKa = 11.15SSTPRR272 pKa = 11.84GGPLDD277 pKa = 3.53RR278 pKa = 11.84AVTRR282 pKa = 11.84FRR284 pKa = 11.84DD285 pKa = 3.77LYY287 pKa = 10.26NRR289 pKa = 11.84RR290 pKa = 11.84VQQVRR295 pKa = 11.84VSDD298 pKa = 3.99PEE300 pKa = 4.06QFLSAPARR308 pKa = 11.84AVTFEE313 pKa = 4.09NPAFDD318 pKa = 4.28SGDD321 pKa = 2.85ISLIFEE327 pKa = 3.99QDD329 pKa = 3.3VQAVQATPDD338 pKa = 4.15PDD340 pKa = 3.61FMDD343 pKa = 5.15IIRR346 pKa = 11.84LGRR349 pKa = 11.84PRR351 pKa = 11.84LSEE354 pKa = 3.59TADD357 pKa = 3.25RR358 pKa = 11.84YY359 pKa = 10.52VRR361 pKa = 11.84VSRR364 pKa = 11.84LGQRR368 pKa = 11.84GTIRR372 pKa = 11.84TRR374 pKa = 11.84SGLQIGGRR382 pKa = 11.84VHH384 pKa = 7.08YY385 pKa = 8.91YY386 pKa = 10.22TDD388 pKa = 4.32LSPIPRR394 pKa = 11.84DD395 pKa = 3.32IEE397 pKa = 4.19LSTLGEE403 pKa = 4.16ASGEE407 pKa = 3.88ADD409 pKa = 3.43LVDD412 pKa = 5.22GIGEE416 pKa = 4.27SSVIDD421 pKa = 3.81AGSVADD427 pKa = 3.94EE428 pKa = 4.43SMLIGEE434 pKa = 4.8GLGAPLDD441 pKa = 4.32DD442 pKa = 5.92LDD444 pKa = 5.63LISDD448 pKa = 4.4GSSVDD453 pKa = 4.12FSSTRR458 pKa = 11.84LEE460 pKa = 4.16FSFLTGSSNITLPEE474 pKa = 3.58QVTNVFGPGLYY485 pKa = 10.49VPDD488 pKa = 4.51IGTGTHH494 pKa = 7.08IIHH497 pKa = 6.3PQVVGEE503 pKa = 4.41VEE505 pKa = 4.72LEE507 pKa = 3.95DD508 pKa = 4.12SFPRR512 pKa = 11.84DD513 pKa = 3.67LFPGTEE519 pKa = 3.58IVFIPEE525 pKa = 3.98DD526 pKa = 3.82SIDD529 pKa = 4.14FYY531 pKa = 11.3LHH533 pKa = 6.17PSLRR537 pKa = 11.84RR538 pKa = 11.84RR539 pKa = 11.84KK540 pKa = 8.82RR541 pKa = 11.84KK542 pKa = 9.74YY543 pKa = 9.81FVRR546 pKa = 4.99
MM1 pKa = 7.25VSATRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VKK10 pKa = 10.34RR11 pKa = 11.84DD12 pKa = 3.07SASNLYY18 pKa = 9.65RR19 pKa = 11.84QCQVTGNCPPDD30 pKa = 3.41VANKK34 pKa = 10.19IEE36 pKa = 4.27GNTLADD42 pKa = 3.59KK43 pKa = 10.53LLKK46 pKa = 10.12IFSSIIYY53 pKa = 10.17LGGLGIGTGRR63 pKa = 11.84GTGEE67 pKa = 4.0TYY69 pKa = 10.56GYY71 pKa = 11.24GPINPGGGRR80 pKa = 11.84ITGTGTVMRR89 pKa = 11.84PGVVVEE95 pKa = 4.83PIGPAEE101 pKa = 4.09IVPVDD106 pKa = 3.64SVGPGEE112 pKa = 4.24SSIVPLLEE120 pKa = 4.05ATPDD124 pKa = 3.45VPINGGPEE132 pKa = 3.87VPPAGPDD139 pKa = 3.14TSTVDD144 pKa = 3.23VTSNIDD150 pKa = 3.32PVSDD154 pKa = 3.75VNVSGGTTITNTDD167 pKa = 3.28SAVIDD172 pKa = 3.9VQPAPSGPRR181 pKa = 11.84RR182 pKa = 11.84VTVTRR187 pKa = 11.84SEE189 pKa = 4.53HH190 pKa = 6.01NNPAYY195 pKa = 10.68VSVSAPPQGLGEE207 pKa = 4.39GGGFLLAGSEE217 pKa = 4.26SSGLIASTHH226 pKa = 5.62EE227 pKa = 3.93LDD229 pKa = 3.42AAVIIGGRR237 pKa = 11.84PPPEE241 pKa = 4.2TIFEE245 pKa = 4.98DD246 pKa = 3.55IEE248 pKa = 5.74LDD250 pKa = 3.65TFSATGGRR258 pKa = 11.84SEE260 pKa = 4.46FDD262 pKa = 2.76IEE264 pKa = 4.33EE265 pKa = 4.99PYY267 pKa = 11.15SSTPRR272 pKa = 11.84GGPLDD277 pKa = 3.53RR278 pKa = 11.84AVTRR282 pKa = 11.84FRR284 pKa = 11.84DD285 pKa = 3.77LYY287 pKa = 10.26NRR289 pKa = 11.84RR290 pKa = 11.84VQQVRR295 pKa = 11.84VSDD298 pKa = 3.99PEE300 pKa = 4.06QFLSAPARR308 pKa = 11.84AVTFEE313 pKa = 4.09NPAFDD318 pKa = 4.28SGDD321 pKa = 2.85ISLIFEE327 pKa = 3.99QDD329 pKa = 3.3VQAVQATPDD338 pKa = 4.15PDD340 pKa = 3.61FMDD343 pKa = 5.15IIRR346 pKa = 11.84LGRR349 pKa = 11.84PRR351 pKa = 11.84LSEE354 pKa = 3.59TADD357 pKa = 3.25RR358 pKa = 11.84YY359 pKa = 10.52VRR361 pKa = 11.84VSRR364 pKa = 11.84LGQRR368 pKa = 11.84GTIRR372 pKa = 11.84TRR374 pKa = 11.84SGLQIGGRR382 pKa = 11.84VHH384 pKa = 7.08YY385 pKa = 8.91YY386 pKa = 10.22TDD388 pKa = 4.32LSPIPRR394 pKa = 11.84DD395 pKa = 3.32IEE397 pKa = 4.19LSTLGEE403 pKa = 4.16ASGEE407 pKa = 3.88ADD409 pKa = 3.43LVDD412 pKa = 5.22GIGEE416 pKa = 4.27SSVIDD421 pKa = 3.81AGSVADD427 pKa = 3.94EE428 pKa = 4.43SMLIGEE434 pKa = 4.8GLGAPLDD441 pKa = 4.32DD442 pKa = 5.92LDD444 pKa = 5.63LISDD448 pKa = 4.4GSSVDD453 pKa = 4.12FSSTRR458 pKa = 11.84LEE460 pKa = 4.16FSFLTGSSNITLPEE474 pKa = 3.58QVTNVFGPGLYY485 pKa = 10.49VPDD488 pKa = 4.51IGTGTHH494 pKa = 7.08IIHH497 pKa = 6.3PQVVGEE503 pKa = 4.41VEE505 pKa = 4.72LEE507 pKa = 3.95DD508 pKa = 4.12SFPRR512 pKa = 11.84DD513 pKa = 3.67LFPGTEE519 pKa = 3.58IVFIPEE525 pKa = 3.98DD526 pKa = 3.82SIDD529 pKa = 4.14FYY531 pKa = 11.3LHH533 pKa = 6.17PSLRR537 pKa = 11.84RR538 pKa = 11.84RR539 pKa = 11.84KK540 pKa = 8.82RR541 pKa = 11.84KK542 pKa = 9.74YY543 pKa = 9.81FVRR546 pKa = 4.99
Molecular weight: 58.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C8AY90|C8AY90_RNPV1 Protein E7 OS=Rattus norvegicus papillomavirus 1 (isolate Rat/Germany/Schulz/2009) OX=664730 GN=E7 PE=3 SV=1
MM1 pKa = 7.03TLKK4 pKa = 10.83GYY6 pKa = 7.94TLLDD10 pKa = 3.44LCRR13 pKa = 11.84YY14 pKa = 9.18YY15 pKa = 10.94DD16 pKa = 3.59VSPTSFPLDD25 pKa = 3.56CTFCDD30 pKa = 4.43CVLDD34 pKa = 4.29DD35 pKa = 4.03VDD37 pKa = 4.1RR38 pKa = 11.84ASFAASKK45 pKa = 10.65LKK47 pKa = 10.8VVVRR51 pKa = 11.84NLQYY55 pKa = 10.91KK56 pKa = 7.81GACIKK61 pKa = 10.37CRR63 pKa = 11.84RR64 pKa = 11.84ALAAEE69 pKa = 4.39EE70 pKa = 4.07KK71 pKa = 10.28FKK73 pKa = 11.44YY74 pKa = 9.54LVCVGEE80 pKa = 4.41ADD82 pKa = 4.71LVEE85 pKa = 4.42AMCGTGIVHH94 pKa = 6.67CSVRR98 pKa = 11.84CVSCLALLSASEE110 pKa = 4.1KK111 pKa = 10.62LVAKK115 pKa = 9.83GGCQPFYY122 pKa = 10.77LVRR125 pKa = 11.84HH126 pKa = 6.3LWRR129 pKa = 11.84AQCRR133 pKa = 11.84HH134 pKa = 5.89CRR136 pKa = 11.84TPGCC140 pKa = 4.15
MM1 pKa = 7.03TLKK4 pKa = 10.83GYY6 pKa = 7.94TLLDD10 pKa = 3.44LCRR13 pKa = 11.84YY14 pKa = 9.18YY15 pKa = 10.94DD16 pKa = 3.59VSPTSFPLDD25 pKa = 3.56CTFCDD30 pKa = 4.43CVLDD34 pKa = 4.29DD35 pKa = 4.03VDD37 pKa = 4.1RR38 pKa = 11.84ASFAASKK45 pKa = 10.65LKK47 pKa = 10.8VVVRR51 pKa = 11.84NLQYY55 pKa = 10.91KK56 pKa = 7.81GACIKK61 pKa = 10.37CRR63 pKa = 11.84RR64 pKa = 11.84ALAAEE69 pKa = 4.39EE70 pKa = 4.07KK71 pKa = 10.28FKK73 pKa = 11.44YY74 pKa = 9.54LVCVGEE80 pKa = 4.41ADD82 pKa = 4.71LVEE85 pKa = 4.42AMCGTGIVHH94 pKa = 6.67CSVRR98 pKa = 11.84CVSCLALLSASEE110 pKa = 4.1KK111 pKa = 10.62LVAKK115 pKa = 9.83GGCQPFYY122 pKa = 10.77LVRR125 pKa = 11.84HH126 pKa = 6.3LWRR129 pKa = 11.84AQCRR133 pKa = 11.84HH134 pKa = 5.89CRR136 pKa = 11.84TPGCC140 pKa = 4.15
Molecular weight: 15.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2289 |
101 |
604 |
381.5 |
42.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.417 ± 0.599 | 2.359 ± 0.822 |
6.815 ± 0.246 | 5.505 ± 0.694 |
4.543 ± 0.564 | 8.17 ± 0.884 |
2.053 ± 0.339 | 3.976 ± 0.853 |
4.281 ± 0.873 | 8.475 ± 0.553 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.529 ± 0.238 | 3.844 ± 0.749 |
6.291 ± 0.914 | 4.063 ± 0.555 |
6.772 ± 0.456 | 7.165 ± 0.686 |
7.252 ± 0.501 | 7.252 ± 0.613 |
1.18 ± 0.351 | 3.058 ± 0.313 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
