
Pseudorhodobacter sp. E13
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pseudorhodobacter; unclassified Pseudorhodobacter
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
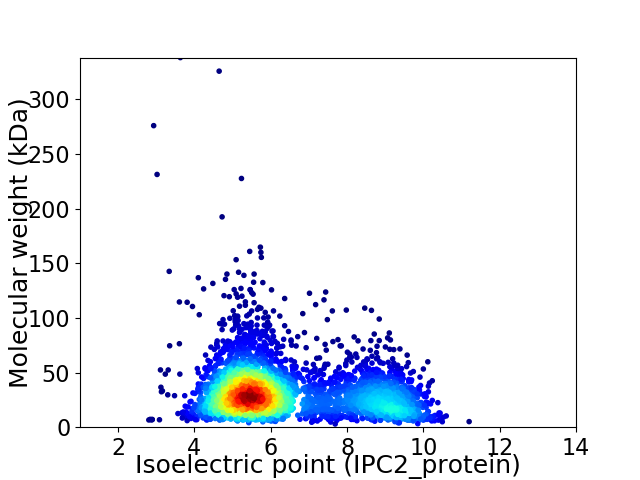
Virtual 2D-PAGE plot for 3709 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
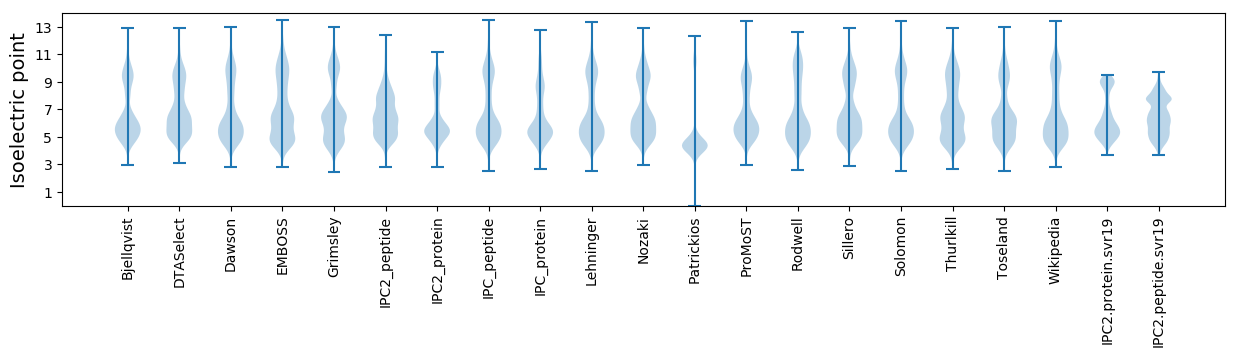
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S1AMU9|A0A3S1AMU9_9RHOB Methyltransferase OS=Pseudorhodobacter sp. E13 OX=2487931 GN=EGN72_11845 PE=3 SV=1
MM1 pKa = 6.52VTNTNTNPGAQTNQTITASSGTDD24 pKa = 3.05TLTGGLGNDD33 pKa = 4.76TINGLAGNDD42 pKa = 3.7FLRR45 pKa = 11.84GDD47 pKa = 4.13GAVQGAWHH55 pKa = 6.7YY56 pKa = 7.58EE57 pKa = 3.79TFNYY61 pKa = 10.11NFSSANGQAFTPAAFTTATRR81 pKa = 11.84TGSGYY86 pKa = 8.81VTDD89 pKa = 4.47FDD91 pKa = 4.53EE92 pKa = 5.88GGITNTMRR100 pKa = 11.84GVAASTNPEE109 pKa = 4.15DD110 pKa = 3.83FGVVYY115 pKa = 10.29TSTLNVTTGGTYY127 pKa = 10.75RR128 pKa = 11.84LTTSSDD134 pKa = 3.42DD135 pKa = 3.49GSTVQIFNSAGVPLNFSNQTGGTLNYY161 pKa = 10.06LNNDD165 pKa = 3.62FHH167 pKa = 8.64QSTTTRR173 pKa = 11.84WGDD176 pKa = 3.3VVLAPGQTYY185 pKa = 8.59TIQIRR190 pKa = 11.84YY191 pKa = 7.76WEE193 pKa = 4.21NLGADD198 pKa = 3.78TLAATINGPDD208 pKa = 3.38TGGATQNLLTSPMLGLPPGPSYY230 pKa = 11.37SVTGTAMGVEE240 pKa = 4.6GNDD243 pKa = 3.59VLDD246 pKa = 4.55GGAGNDD252 pKa = 3.84TIYY255 pKa = 11.3GDD257 pKa = 4.05GGNDD261 pKa = 3.49SLLGGADD268 pKa = 3.39NDD270 pKa = 4.11LLYY273 pKa = 11.13GGTGNDD279 pKa = 3.37TLFGGTGNDD288 pKa = 3.31TLYY291 pKa = 11.33GDD293 pKa = 5.17DD294 pKa = 5.67SSDD297 pKa = 4.13SLNGDD302 pKa = 3.85DD303 pKa = 5.81GTDD306 pKa = 3.5LLYY309 pKa = 11.19GGAGNDD315 pKa = 3.89TLNGGAGADD324 pKa = 3.65TLYY327 pKa = 11.32GDD329 pKa = 5.41DD330 pKa = 4.93GNDD333 pKa = 3.55SLRR336 pKa = 11.84GDD338 pKa = 4.03ADD340 pKa = 3.6NDD342 pKa = 3.99TLYY345 pKa = 11.28GGAGLDD351 pKa = 3.89TLNGDD356 pKa = 4.52DD357 pKa = 5.82GNDD360 pKa = 3.52ALYY363 pKa = 10.92GGADD367 pKa = 3.5ADD369 pKa = 3.86QLFGGLGSDD378 pKa = 3.84SLFGGTGADD387 pKa = 3.18TLYY390 pKa = 11.08GGDD393 pKa = 4.06GADD396 pKa = 4.22SLSGDD401 pKa = 4.6DD402 pKa = 5.35GNDD405 pKa = 3.32SLFGGLGDD413 pKa = 3.96DD414 pKa = 4.07TLLGGLGNDD423 pKa = 3.76TLYY426 pKa = 11.43GDD428 pKa = 5.45DD429 pKa = 5.76GNDD432 pKa = 3.73SLNGDD437 pKa = 4.3AGNDD441 pKa = 3.69SLFGGAGNDD450 pKa = 3.84TLNGGAGNNTLTGGAGNDD468 pKa = 3.05RR469 pKa = 11.84FIYY472 pKa = 9.98TIGSNLTISDD482 pKa = 4.47LNLGNSGGILDD493 pKa = 5.19GDD495 pKa = 3.68QTNNDD500 pKa = 4.04FLDD503 pKa = 4.06LSAFYY508 pKa = 10.85TNLNEE513 pKa = 4.89LRR515 pKa = 11.84DD516 pKa = 4.19DD517 pKa = 4.29FSDD520 pKa = 5.28DD521 pKa = 4.17GILNQSVGDD530 pKa = 3.98YY531 pKa = 11.08SDD533 pKa = 3.36NTALGGSIILTGITRR548 pKa = 11.84TDD550 pKa = 3.25LTTDD554 pKa = 4.23NINVACFTAGTQIEE568 pKa = 4.67TADD571 pKa = 3.89GPALIEE577 pKa = 4.25SLTAGTLIRR586 pKa = 11.84TLDD589 pKa = 3.56HH590 pKa = 6.75GLQPLRR596 pKa = 11.84AVLTRR601 pKa = 11.84SVPGDD606 pKa = 3.28GRR608 pKa = 11.84FAPIRR613 pKa = 11.84FHH615 pKa = 7.66AGALGNHH622 pKa = 6.43RR623 pKa = 11.84AFSTSPAHH631 pKa = 7.21RR632 pKa = 11.84MLIADD637 pKa = 4.09WRR639 pKa = 11.84AEE641 pKa = 3.86LLFGEE646 pKa = 4.54SEE648 pKa = 4.38VLISARR654 pKa = 11.84ALVNDD659 pKa = 4.08CTITRR664 pKa = 11.84APCDD668 pKa = 2.99RR669 pKa = 11.84VTYY672 pKa = 10.01YY673 pKa = 10.94HH674 pKa = 7.17LLLDD678 pKa = 3.31RR679 pKa = 11.84HH680 pKa = 6.18EE681 pKa = 5.06IIFAEE686 pKa = 5.6GIATEE691 pKa = 4.88SYY693 pKa = 9.33QHH695 pKa = 6.59GAFADD700 pKa = 3.98DD701 pKa = 4.35PGVEE705 pKa = 4.25AEE707 pKa = 4.98LDD709 pKa = 3.54ALFPGLLPAPSDD721 pKa = 3.52SARR724 pKa = 11.84PSLRR728 pKa = 11.84GYY730 pKa = 9.16EE731 pKa = 3.98AAALLSLPQRR741 pKa = 11.84RR742 pKa = 11.84AAA744 pKa = 4.17
MM1 pKa = 6.52VTNTNTNPGAQTNQTITASSGTDD24 pKa = 3.05TLTGGLGNDD33 pKa = 4.76TINGLAGNDD42 pKa = 3.7FLRR45 pKa = 11.84GDD47 pKa = 4.13GAVQGAWHH55 pKa = 6.7YY56 pKa = 7.58EE57 pKa = 3.79TFNYY61 pKa = 10.11NFSSANGQAFTPAAFTTATRR81 pKa = 11.84TGSGYY86 pKa = 8.81VTDD89 pKa = 4.47FDD91 pKa = 4.53EE92 pKa = 5.88GGITNTMRR100 pKa = 11.84GVAASTNPEE109 pKa = 4.15DD110 pKa = 3.83FGVVYY115 pKa = 10.29TSTLNVTTGGTYY127 pKa = 10.75RR128 pKa = 11.84LTTSSDD134 pKa = 3.42DD135 pKa = 3.49GSTVQIFNSAGVPLNFSNQTGGTLNYY161 pKa = 10.06LNNDD165 pKa = 3.62FHH167 pKa = 8.64QSTTTRR173 pKa = 11.84WGDD176 pKa = 3.3VVLAPGQTYY185 pKa = 8.59TIQIRR190 pKa = 11.84YY191 pKa = 7.76WEE193 pKa = 4.21NLGADD198 pKa = 3.78TLAATINGPDD208 pKa = 3.38TGGATQNLLTSPMLGLPPGPSYY230 pKa = 11.37SVTGTAMGVEE240 pKa = 4.6GNDD243 pKa = 3.59VLDD246 pKa = 4.55GGAGNDD252 pKa = 3.84TIYY255 pKa = 11.3GDD257 pKa = 4.05GGNDD261 pKa = 3.49SLLGGADD268 pKa = 3.39NDD270 pKa = 4.11LLYY273 pKa = 11.13GGTGNDD279 pKa = 3.37TLFGGTGNDD288 pKa = 3.31TLYY291 pKa = 11.33GDD293 pKa = 5.17DD294 pKa = 5.67SSDD297 pKa = 4.13SLNGDD302 pKa = 3.85DD303 pKa = 5.81GTDD306 pKa = 3.5LLYY309 pKa = 11.19GGAGNDD315 pKa = 3.89TLNGGAGADD324 pKa = 3.65TLYY327 pKa = 11.32GDD329 pKa = 5.41DD330 pKa = 4.93GNDD333 pKa = 3.55SLRR336 pKa = 11.84GDD338 pKa = 4.03ADD340 pKa = 3.6NDD342 pKa = 3.99TLYY345 pKa = 11.28GGAGLDD351 pKa = 3.89TLNGDD356 pKa = 4.52DD357 pKa = 5.82GNDD360 pKa = 3.52ALYY363 pKa = 10.92GGADD367 pKa = 3.5ADD369 pKa = 3.86QLFGGLGSDD378 pKa = 3.84SLFGGTGADD387 pKa = 3.18TLYY390 pKa = 11.08GGDD393 pKa = 4.06GADD396 pKa = 4.22SLSGDD401 pKa = 4.6DD402 pKa = 5.35GNDD405 pKa = 3.32SLFGGLGDD413 pKa = 3.96DD414 pKa = 4.07TLLGGLGNDD423 pKa = 3.76TLYY426 pKa = 11.43GDD428 pKa = 5.45DD429 pKa = 5.76GNDD432 pKa = 3.73SLNGDD437 pKa = 4.3AGNDD441 pKa = 3.69SLFGGAGNDD450 pKa = 3.84TLNGGAGNNTLTGGAGNDD468 pKa = 3.05RR469 pKa = 11.84FIYY472 pKa = 9.98TIGSNLTISDD482 pKa = 4.47LNLGNSGGILDD493 pKa = 5.19GDD495 pKa = 3.68QTNNDD500 pKa = 4.04FLDD503 pKa = 4.06LSAFYY508 pKa = 10.85TNLNEE513 pKa = 4.89LRR515 pKa = 11.84DD516 pKa = 4.19DD517 pKa = 4.29FSDD520 pKa = 5.28DD521 pKa = 4.17GILNQSVGDD530 pKa = 3.98YY531 pKa = 11.08SDD533 pKa = 3.36NTALGGSIILTGITRR548 pKa = 11.84TDD550 pKa = 3.25LTTDD554 pKa = 4.23NINVACFTAGTQIEE568 pKa = 4.67TADD571 pKa = 3.89GPALIEE577 pKa = 4.25SLTAGTLIRR586 pKa = 11.84TLDD589 pKa = 3.56HH590 pKa = 6.75GLQPLRR596 pKa = 11.84AVLTRR601 pKa = 11.84SVPGDD606 pKa = 3.28GRR608 pKa = 11.84FAPIRR613 pKa = 11.84FHH615 pKa = 7.66AGALGNHH622 pKa = 6.43RR623 pKa = 11.84AFSTSPAHH631 pKa = 7.21RR632 pKa = 11.84MLIADD637 pKa = 4.09WRR639 pKa = 11.84AEE641 pKa = 3.86LLFGEE646 pKa = 4.54SEE648 pKa = 4.38VLISARR654 pKa = 11.84ALVNDD659 pKa = 4.08CTITRR664 pKa = 11.84APCDD668 pKa = 2.99RR669 pKa = 11.84VTYY672 pKa = 10.01YY673 pKa = 10.94HH674 pKa = 7.17LLLDD678 pKa = 3.31RR679 pKa = 11.84HH680 pKa = 6.18EE681 pKa = 5.06IIFAEE686 pKa = 5.6GIATEE691 pKa = 4.88SYY693 pKa = 9.33QHH695 pKa = 6.59GAFADD700 pKa = 3.98DD701 pKa = 4.35PGVEE705 pKa = 4.25AEE707 pKa = 4.98LDD709 pKa = 3.54ALFPGLLPAPSDD721 pKa = 3.52SARR724 pKa = 11.84PSLRR728 pKa = 11.84GYY730 pKa = 9.16EE731 pKa = 3.98AAALLSLPQRR741 pKa = 11.84RR742 pKa = 11.84AAA744 pKa = 4.17
Molecular weight: 76.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S0YYZ7|A0A3S0YYZ7_9RHOB DUF1501 domain-containing protein OS=Pseudorhodobacter sp. E13 OX=2487931 GN=EGN72_07320 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.33GGRR28 pKa = 11.84IVLNNRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.11GRR39 pKa = 11.84KK40 pKa = 8.82KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.33GGRR28 pKa = 11.84IVLNNRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.11GRR39 pKa = 11.84KK40 pKa = 8.82KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1156015 |
25 |
3543 |
311.7 |
33.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.292 ± 0.061 | 0.887 ± 0.013 |
5.736 ± 0.046 | 5.418 ± 0.037 |
3.716 ± 0.028 | 8.787 ± 0.053 |
2.005 ± 0.02 | 5.008 ± 0.026 |
3.342 ± 0.038 | 10.282 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.782 ± 0.024 | 2.582 ± 0.026 |
5.209 ± 0.037 | 3.319 ± 0.019 |
6.506 ± 0.044 | 4.99 ± 0.031 |
5.492 ± 0.051 | 7.067 ± 0.034 |
1.399 ± 0.017 | 2.182 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
