
Human papillomavirus sp.
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; Human papillomavirus types
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
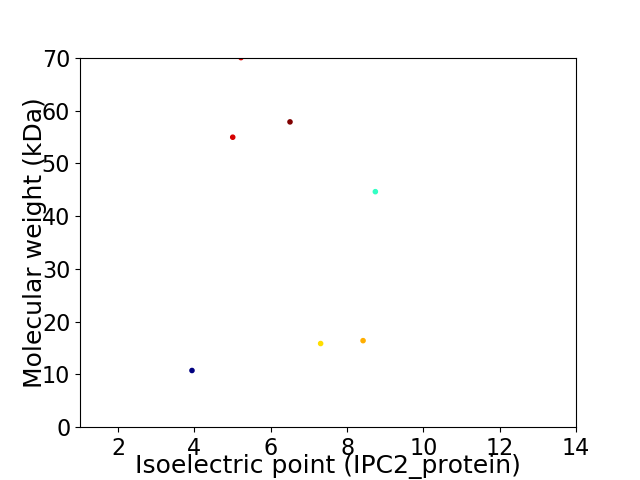
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
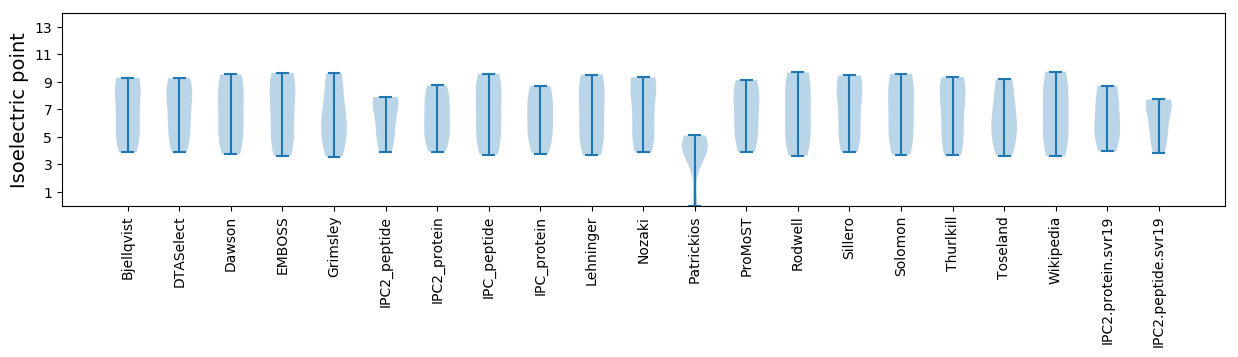
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2AMJ6|A0A2D2AMJ6_9PAPI E4 OS=Human papillomavirus sp. OX=192971 GN=E4 PE=4 SV=1
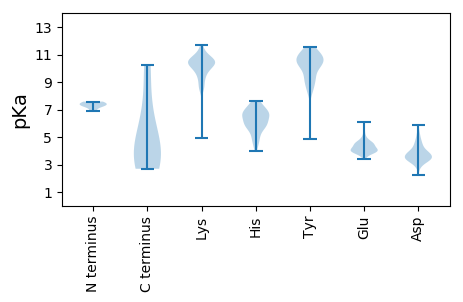
MM1 pKa = 7.29RR2 pKa = 11.84QVSNFSSINNILQEE16 pKa = 4.32EE17 pKa = 4.83EE18 pKa = 3.96PTIIDD23 pKa = 3.83LFCYY27 pKa = 10.52EE28 pKa = 4.46EE29 pKa = 4.2VALSDD34 pKa = 4.02EE35 pKa = 4.63EE36 pKa = 5.09EE37 pKa = 4.38EE38 pKa = 4.38SQQVQQAYY46 pKa = 10.11LVTVQCAHH54 pKa = 6.98CLNLVTFNYY63 pKa = 9.46WADD66 pKa = 3.51LPAIRR71 pKa = 11.84EE72 pKa = 4.15LQQLLFDD79 pKa = 4.55CVHH82 pKa = 6.58FVCEE86 pKa = 3.99SCAAEE91 pKa = 4.09LQQ93 pKa = 3.69
MM1 pKa = 7.29RR2 pKa = 11.84QVSNFSSINNILQEE16 pKa = 4.32EE17 pKa = 4.83EE18 pKa = 3.96PTIIDD23 pKa = 3.83LFCYY27 pKa = 10.52EE28 pKa = 4.46EE29 pKa = 4.2VALSDD34 pKa = 4.02EE35 pKa = 4.63EE36 pKa = 5.09EE37 pKa = 4.38EE38 pKa = 4.38SQQVQQAYY46 pKa = 10.11LVTVQCAHH54 pKa = 6.98CLNLVTFNYY63 pKa = 9.46WADD66 pKa = 3.51LPAIRR71 pKa = 11.84EE72 pKa = 4.15LQQLLFDD79 pKa = 4.55CVHH82 pKa = 6.58FVCEE86 pKa = 3.99SCAAEE91 pKa = 4.09LQQ93 pKa = 3.69
Molecular weight: 10.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2AMG1|A0A2D2AMG1_9PAPI Minor capsid protein L2 OS=Human papillomavirus sp. OX=192971 GN=L2 PE=3 SV=1
MM1 pKa = 7.52EE2 pKa = 5.36NLVQRR7 pKa = 11.84LEE9 pKa = 4.16SLQEE13 pKa = 3.95RR14 pKa = 11.84LLSLYY19 pKa = 10.04EE20 pKa = 3.82QDD22 pKa = 4.89SNDD25 pKa = 3.17IQDD28 pKa = 5.09QITHH32 pKa = 5.15WTLVKK37 pKa = 10.26QEE39 pKa = 3.9QLIYY43 pKa = 10.52HH44 pKa = 6.17YY45 pKa = 10.82ARR47 pKa = 11.84KK48 pKa = 10.11NGIRR52 pKa = 11.84RR53 pKa = 11.84LGMQSLPTLAASEE66 pKa = 4.47TKK68 pKa = 10.25AKK70 pKa = 10.11QAIEE74 pKa = 4.7MILQLQSLANSPFGLEE90 pKa = 3.82SWSLHH95 pKa = 4.55DD96 pKa = 3.72TSRR99 pKa = 11.84EE100 pKa = 3.87RR101 pKa = 11.84YY102 pKa = 4.85TTEE105 pKa = 4.17PEE107 pKa = 3.88NTFKK111 pKa = 10.9KK112 pKa = 10.29QPQTLLITFDD122 pKa = 3.62NDD124 pKa = 2.99RR125 pKa = 11.84DD126 pKa = 3.72NSVEE130 pKa = 4.14HH131 pKa = 5.71TVWTYY136 pKa = 11.26VYY138 pKa = 9.26YY139 pKa = 10.95QNGDD143 pKa = 4.27DD144 pKa = 3.39IWHH147 pKa = 6.68KK148 pKa = 10.71EE149 pKa = 3.85EE150 pKa = 4.9SSVDD154 pKa = 3.25EE155 pKa = 4.18KK156 pKa = 11.45GIYY159 pKa = 8.24FLKK162 pKa = 10.92YY163 pKa = 9.26GVEE166 pKa = 3.45KK167 pKa = 10.28HH168 pKa = 6.29YY169 pKa = 10.95YY170 pKa = 8.32VQFANEE176 pKa = 3.7ATRR179 pKa = 11.84YY180 pKa = 8.36SKK182 pKa = 10.7KK183 pKa = 10.78GEE185 pKa = 3.83YY186 pKa = 8.75TVQFKK191 pKa = 9.33NQRR194 pKa = 11.84HH195 pKa = 5.73SYY197 pKa = 8.6NVSSVSSTSGSPGSPDD213 pKa = 3.02STKK216 pKa = 11.28ANASGSNTRR225 pKa = 11.84STSEE229 pKa = 3.89EE230 pKa = 3.99GPEE233 pKa = 3.43RR234 pKa = 11.84SIRR237 pKa = 11.84SRR239 pKa = 11.84TFGQRR244 pKa = 11.84PSTSPRR250 pKa = 11.84VSSRR254 pKa = 11.84RR255 pKa = 11.84GGQQGKK261 pKa = 8.95PDD263 pKa = 3.65TGADD267 pKa = 3.78SDD269 pKa = 4.52SGLAPPSPEE278 pKa = 3.74EE279 pKa = 3.87VGSRR283 pKa = 11.84TTQPTRR289 pKa = 11.84RR290 pKa = 11.84GQSRR294 pKa = 11.84LRR296 pKa = 11.84VLLQEE301 pKa = 4.48ARR303 pKa = 11.84DD304 pKa = 3.86PLVLCLKK311 pKa = 10.45GGPNQLKK318 pKa = 9.96CLRR321 pKa = 11.84YY322 pKa = 8.85RR323 pKa = 11.84LKK325 pKa = 10.55KK326 pKa = 9.25QHH328 pKa = 6.59HH329 pKa = 6.29KK330 pKa = 10.8LFTKK334 pKa = 10.47ISTTWHH340 pKa = 5.71WVHH343 pKa = 5.7NTSTDD348 pKa = 3.25RR349 pKa = 11.84VGNARR354 pKa = 11.84MLIQFVNEE362 pKa = 3.87DD363 pKa = 3.19QRR365 pKa = 11.84NRR367 pKa = 11.84FLDD370 pKa = 4.33EE371 pKa = 4.58IIVPKK376 pKa = 10.47DD377 pKa = 2.92IIVYY381 pKa = 9.21RR382 pKa = 11.84GYY384 pKa = 10.89FRR386 pKa = 11.84GYY388 pKa = 10.23
MM1 pKa = 7.52EE2 pKa = 5.36NLVQRR7 pKa = 11.84LEE9 pKa = 4.16SLQEE13 pKa = 3.95RR14 pKa = 11.84LLSLYY19 pKa = 10.04EE20 pKa = 3.82QDD22 pKa = 4.89SNDD25 pKa = 3.17IQDD28 pKa = 5.09QITHH32 pKa = 5.15WTLVKK37 pKa = 10.26QEE39 pKa = 3.9QLIYY43 pKa = 10.52HH44 pKa = 6.17YY45 pKa = 10.82ARR47 pKa = 11.84KK48 pKa = 10.11NGIRR52 pKa = 11.84RR53 pKa = 11.84LGMQSLPTLAASEE66 pKa = 4.47TKK68 pKa = 10.25AKK70 pKa = 10.11QAIEE74 pKa = 4.7MILQLQSLANSPFGLEE90 pKa = 3.82SWSLHH95 pKa = 4.55DD96 pKa = 3.72TSRR99 pKa = 11.84EE100 pKa = 3.87RR101 pKa = 11.84YY102 pKa = 4.85TTEE105 pKa = 4.17PEE107 pKa = 3.88NTFKK111 pKa = 10.9KK112 pKa = 10.29QPQTLLITFDD122 pKa = 3.62NDD124 pKa = 2.99RR125 pKa = 11.84DD126 pKa = 3.72NSVEE130 pKa = 4.14HH131 pKa = 5.71TVWTYY136 pKa = 11.26VYY138 pKa = 9.26YY139 pKa = 10.95QNGDD143 pKa = 4.27DD144 pKa = 3.39IWHH147 pKa = 6.68KK148 pKa = 10.71EE149 pKa = 3.85EE150 pKa = 4.9SSVDD154 pKa = 3.25EE155 pKa = 4.18KK156 pKa = 11.45GIYY159 pKa = 8.24FLKK162 pKa = 10.92YY163 pKa = 9.26GVEE166 pKa = 3.45KK167 pKa = 10.28HH168 pKa = 6.29YY169 pKa = 10.95YY170 pKa = 8.32VQFANEE176 pKa = 3.7ATRR179 pKa = 11.84YY180 pKa = 8.36SKK182 pKa = 10.7KK183 pKa = 10.78GEE185 pKa = 3.83YY186 pKa = 8.75TVQFKK191 pKa = 9.33NQRR194 pKa = 11.84HH195 pKa = 5.73SYY197 pKa = 8.6NVSSVSSTSGSPGSPDD213 pKa = 3.02STKK216 pKa = 11.28ANASGSNTRR225 pKa = 11.84STSEE229 pKa = 3.89EE230 pKa = 3.99GPEE233 pKa = 3.43RR234 pKa = 11.84SIRR237 pKa = 11.84SRR239 pKa = 11.84TFGQRR244 pKa = 11.84PSTSPRR250 pKa = 11.84VSSRR254 pKa = 11.84RR255 pKa = 11.84GGQQGKK261 pKa = 8.95PDD263 pKa = 3.65TGADD267 pKa = 3.78SDD269 pKa = 4.52SGLAPPSPEE278 pKa = 3.74EE279 pKa = 3.87VGSRR283 pKa = 11.84TTQPTRR289 pKa = 11.84RR290 pKa = 11.84GQSRR294 pKa = 11.84LRR296 pKa = 11.84VLLQEE301 pKa = 4.48ARR303 pKa = 11.84DD304 pKa = 3.86PLVLCLKK311 pKa = 10.45GGPNQLKK318 pKa = 9.96CLRR321 pKa = 11.84YY322 pKa = 8.85RR323 pKa = 11.84LKK325 pKa = 10.55KK326 pKa = 9.25QHH328 pKa = 6.59HH329 pKa = 6.29KK330 pKa = 10.8LFTKK334 pKa = 10.47ISTTWHH340 pKa = 5.71WVHH343 pKa = 5.7NTSTDD348 pKa = 3.25RR349 pKa = 11.84VGNARR354 pKa = 11.84MLIQFVNEE362 pKa = 3.87DD363 pKa = 3.19QRR365 pKa = 11.84NRR367 pKa = 11.84FLDD370 pKa = 4.33EE371 pKa = 4.58IIVPKK376 pKa = 10.47DD377 pKa = 2.92IIVYY381 pKa = 9.21RR382 pKa = 11.84GYY384 pKa = 10.89FRR386 pKa = 11.84GYY388 pKa = 10.23
Molecular weight: 44.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2384 |
93 |
616 |
340.6 |
38.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.076 ± 0.648 | 1.762 ± 0.671 |
5.956 ± 0.331 | 6.669 ± 0.64 |
4.74 ± 0.488 | 5.495 ± 0.882 |
2.181 ± 0.269 | 5.453 ± 0.839 |
5.201 ± 0.77 | 9.48 ± 1.578 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.342 ± 0.264 | 5.034 ± 0.518 |
5.789 ± 1.006 | 4.992 ± 0.819 |
5.956 ± 0.717 | 7.592 ± 0.864 |
6.711 ± 0.733 | 6.04 ± 0.371 |
1.258 ± 0.363 | 3.272 ± 0.417 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
