
Rhodopseudomonas palustris (strain ATCC BAA-98 / CGA009)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Bradyrhizobiaceae; Rhodopseudomonas; Rhodopseudomonas palustris
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
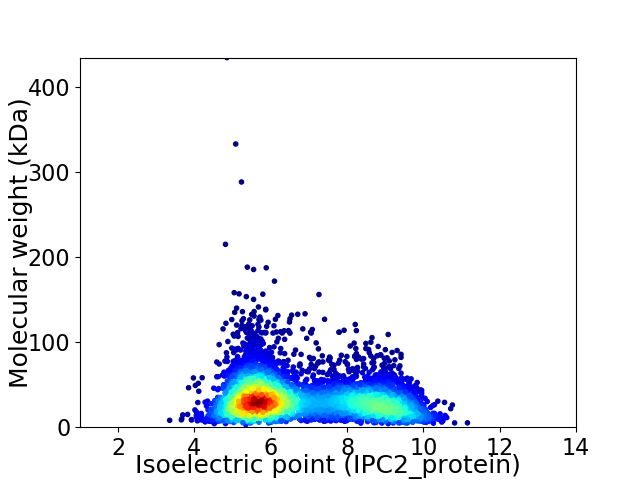
Virtual 2D-PAGE plot for 4811 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
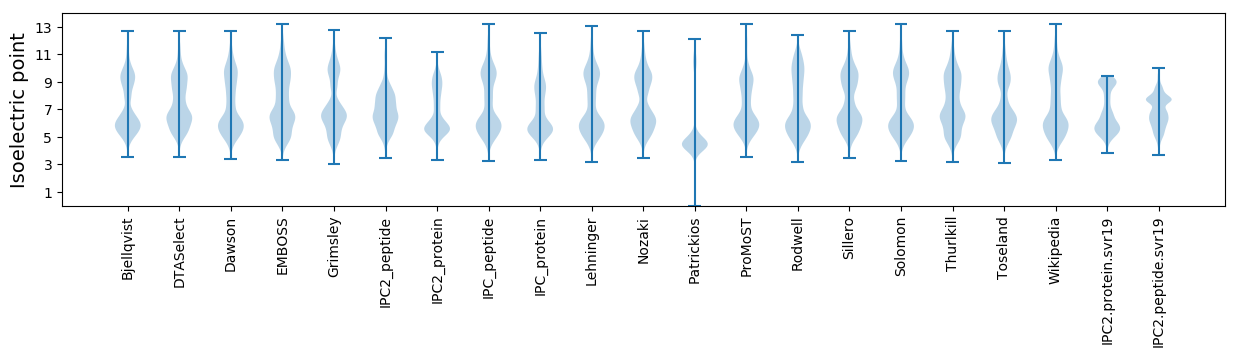
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6NCK3|Q6NCK3_RHOPA Putative sulfate ester transporter ATP-binding component OS=Rhodopseudomonas palustris (strain ATCC BAA-98 / CGA009) OX=258594 GN=AtsC PE=3 SV=1
MM1 pKa = 7.56SLTGALSSAISALNAQSQSLSMISDD26 pKa = 4.62NIANSDD32 pKa = 3.35TTGYY36 pKa = 8.18KK37 pKa = 7.24TTSAMFSQLVTASSSATSYY56 pKa = 11.42ASGGVTVAGRR66 pKa = 11.84ANISQQGLLAATSNATDD83 pKa = 3.37VAIQGSGFFVVSDD96 pKa = 3.61QTSGGTTSYY105 pKa = 9.71TRR107 pKa = 11.84NGAFSIDD114 pKa = 3.33NAGYY118 pKa = 10.01LVNSGYY124 pKa = 10.54YY125 pKa = 9.99LEE127 pKa = 4.73GWRR130 pKa = 11.84TDD132 pKa = 3.12ADD134 pKa = 3.74GKK136 pKa = 10.75IVGDD140 pKa = 3.81ASSATLSPINTNVAATSGSATTKK163 pKa = 8.07TTFAANLPADD173 pKa = 3.99AATGATYY180 pKa = 10.48SSSMTVYY187 pKa = 10.96DD188 pKa = 4.16SLGTANSIQVTWIKK202 pKa = 9.24TASNAWTASFANPTLSSSTSTTTGTTSGSVSLTFNNDD239 pKa = 2.11GSLASTNPSPPTISVSGWTNGAANSTISLDD269 pKa = 3.37LGTVGKK275 pKa = 9.0TNGLTQYY282 pKa = 11.24ASGEE286 pKa = 4.06ATPTINVTSISSDD299 pKa = 2.95GLPYY303 pKa = 10.93GKK305 pKa = 10.14LSSIAVGDD313 pKa = 4.25DD314 pKa = 3.26GTVNATYY321 pKa = 11.24SNGQTIAIYY330 pKa = 9.9KK331 pKa = 9.49IAVATFKK338 pKa = 11.24DD339 pKa = 3.8PDD341 pKa = 3.9GLSVNSYY348 pKa = 10.87GIYY351 pKa = 10.33SVTAASGDD359 pKa = 3.57ASVQTSGSNGAGKK372 pKa = 10.15IYY374 pKa = 10.31GSEE377 pKa = 4.4LEE379 pKa = 4.48SSTTDD384 pKa = 2.93TSSQFASMISAQQAYY399 pKa = 9.56SAASQVITTVNKK411 pKa = 9.81MFDD414 pKa = 3.61TLISAMRR421 pKa = 3.98
MM1 pKa = 7.56SLTGALSSAISALNAQSQSLSMISDD26 pKa = 4.62NIANSDD32 pKa = 3.35TTGYY36 pKa = 8.18KK37 pKa = 7.24TTSAMFSQLVTASSSATSYY56 pKa = 11.42ASGGVTVAGRR66 pKa = 11.84ANISQQGLLAATSNATDD83 pKa = 3.37VAIQGSGFFVVSDD96 pKa = 3.61QTSGGTTSYY105 pKa = 9.71TRR107 pKa = 11.84NGAFSIDD114 pKa = 3.33NAGYY118 pKa = 10.01LVNSGYY124 pKa = 10.54YY125 pKa = 9.99LEE127 pKa = 4.73GWRR130 pKa = 11.84TDD132 pKa = 3.12ADD134 pKa = 3.74GKK136 pKa = 10.75IVGDD140 pKa = 3.81ASSATLSPINTNVAATSGSATTKK163 pKa = 8.07TTFAANLPADD173 pKa = 3.99AATGATYY180 pKa = 10.48SSSMTVYY187 pKa = 10.96DD188 pKa = 4.16SLGTANSIQVTWIKK202 pKa = 9.24TASNAWTASFANPTLSSSTSTTTGTTSGSVSLTFNNDD239 pKa = 2.11GSLASTNPSPPTISVSGWTNGAANSTISLDD269 pKa = 3.37LGTVGKK275 pKa = 9.0TNGLTQYY282 pKa = 11.24ASGEE286 pKa = 4.06ATPTINVTSISSDD299 pKa = 2.95GLPYY303 pKa = 10.93GKK305 pKa = 10.14LSSIAVGDD313 pKa = 4.25DD314 pKa = 3.26GTVNATYY321 pKa = 11.24SNGQTIAIYY330 pKa = 9.9KK331 pKa = 9.49IAVATFKK338 pKa = 11.24DD339 pKa = 3.8PDD341 pKa = 3.9GLSVNSYY348 pKa = 10.87GIYY351 pKa = 10.33SVTAASGDD359 pKa = 3.57ASVQTSGSNGAGKK372 pKa = 10.15IYY374 pKa = 10.31GSEE377 pKa = 4.4LEE379 pKa = 4.48SSTTDD384 pKa = 2.93TSSQFASMISAQQAYY399 pKa = 9.56SAASQVITTVNKK411 pKa = 9.81MFDD414 pKa = 3.61TLISAMRR421 pKa = 3.98
Molecular weight: 42.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6NC61|Q6NC61_RHOPA Cupin_5 domain-containing protein OS=Rhodopseudomonas palustris (strain ATCC BAA-98 / CGA009) OX=258594 GN=RPA0611 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATTGGRR28 pKa = 11.84KK29 pKa = 9.06VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATTGGRR28 pKa = 11.84KK29 pKa = 9.06VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1579866 |
25 |
4335 |
328.4 |
35.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.807 ± 0.046 | 0.85 ± 0.012 |
5.65 ± 0.023 | 5.218 ± 0.033 |
3.641 ± 0.021 | 8.366 ± 0.037 |
1.976 ± 0.018 | 5.263 ± 0.025 |
3.514 ± 0.027 | 10.06 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.341 ± 0.018 | 2.581 ± 0.02 |
5.393 ± 0.029 | 3.282 ± 0.02 |
7.002 ± 0.033 | 5.59 ± 0.025 |
5.465 ± 0.025 | 7.495 ± 0.029 |
1.28 ± 0.014 | 2.224 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
