
Fervidicola ferrireducens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Thermosediminibacterales; Thermosediminibacteraceae; Fervidicola
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
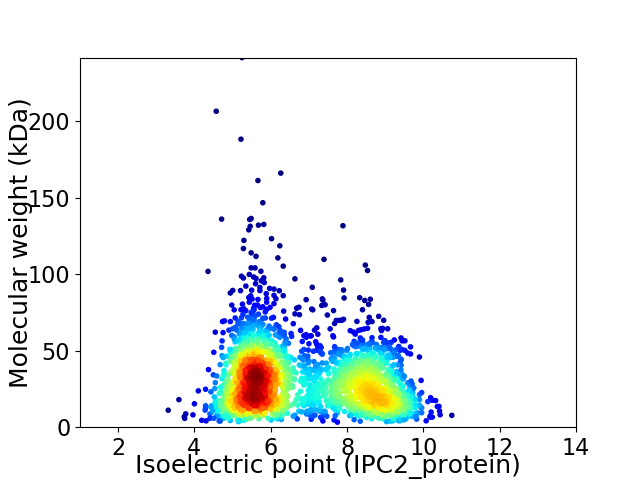
Virtual 2D-PAGE plot for 2421 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
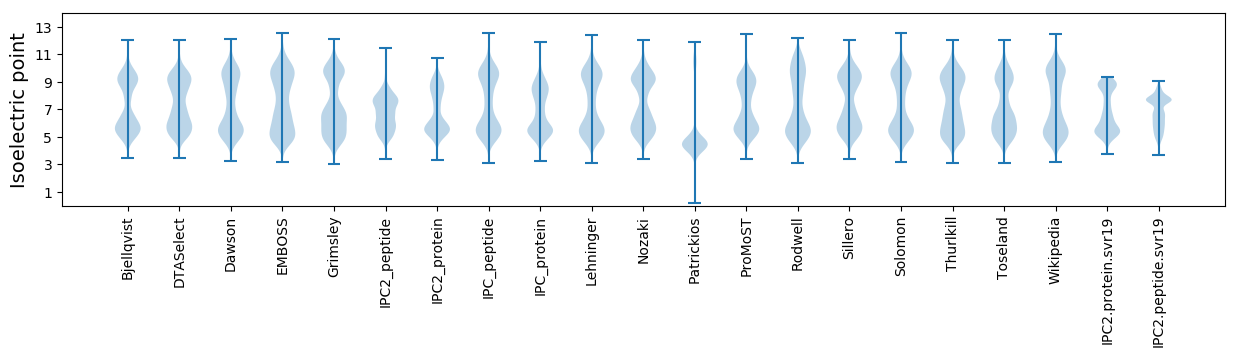
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140L6A5|A0A140L6A5_9FIRM Uncharacterized protein OS=Fervidicola ferrireducens OX=520764 GN=AN618_16680 PE=4 SV=1
MM1 pKa = 7.38KK2 pKa = 10.23KK3 pKa = 9.14WVCTVCGYY11 pKa = 11.14VYY13 pKa = 10.74DD14 pKa = 4.65PDD16 pKa = 5.75KK17 pKa = 11.41GDD19 pKa = 4.02PDD21 pKa = 3.71NGIEE25 pKa = 4.46PGTSFDD31 pKa = 5.6DD32 pKa = 6.0LPDD35 pKa = 4.02DD36 pKa = 3.81WTCPVCGAGKK46 pKa = 10.55DD47 pKa = 3.64MFEE50 pKa = 4.42EE51 pKa = 4.5EE52 pKa = 4.21
MM1 pKa = 7.38KK2 pKa = 10.23KK3 pKa = 9.14WVCTVCGYY11 pKa = 11.14VYY13 pKa = 10.74DD14 pKa = 4.65PDD16 pKa = 5.75KK17 pKa = 11.41GDD19 pKa = 4.02PDD21 pKa = 3.71NGIEE25 pKa = 4.46PGTSFDD31 pKa = 5.6DD32 pKa = 6.0LPDD35 pKa = 4.02DD36 pKa = 3.81WTCPVCGAGKK46 pKa = 10.55DD47 pKa = 3.64MFEE50 pKa = 4.42EE51 pKa = 4.5EE52 pKa = 4.21
Molecular weight: 5.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140L4E1|A0A140L4E1_9FIRM Orotate phosphoribosyltransferase OS=Fervidicola ferrireducens OX=520764 GN=pyrE PE=3 SV=1
MM1 pKa = 7.07GHH3 pKa = 6.96KK4 pKa = 9.87NLKK7 pKa = 9.57QLDD10 pKa = 4.39AIAFKK15 pKa = 10.69PGDD18 pKa = 3.4EE19 pKa = 4.27SGFRR23 pKa = 11.84LCYY26 pKa = 9.39NGSMKK31 pKa = 9.09HH32 pKa = 4.66TNVVRR37 pKa = 11.84VLPDD41 pKa = 3.1KK42 pKa = 10.6RR43 pKa = 11.84QKK45 pKa = 10.27QVLEE49 pKa = 4.47IIGDD53 pKa = 3.87RR54 pKa = 11.84CAALWNAVQYY64 pKa = 10.49RR65 pKa = 11.84CRR67 pKa = 11.84QAFFKK72 pKa = 10.78GDD74 pKa = 3.82SVPSYY79 pKa = 11.1AALCSEE85 pKa = 4.81FKK87 pKa = 10.5EE88 pKa = 4.38HH89 pKa = 6.94PAYY92 pKa = 9.76KK93 pKa = 10.27ALPAHH98 pKa = 7.2IGQEE102 pKa = 4.36VIKK105 pKa = 10.0KK106 pKa = 9.63ARR108 pKa = 11.84KK109 pKa = 8.86AWDD112 pKa = 3.36SYY114 pKa = 9.23FACLRR119 pKa = 11.84LYY121 pKa = 10.7RR122 pKa = 11.84KK123 pKa = 10.33GEE125 pKa = 4.15LKK127 pKa = 10.46EE128 pKa = 4.15PPHH131 pKa = 7.12IPRR134 pKa = 11.84YY135 pKa = 8.09WKK137 pKa = 10.1DD138 pKa = 2.87RR139 pKa = 11.84RR140 pKa = 11.84TGRR143 pKa = 11.84RR144 pKa = 11.84IFRR147 pKa = 11.84VIPVKK152 pKa = 10.88APTSYY157 pKa = 11.47SLDD160 pKa = 3.52ARR162 pKa = 11.84CLSLTLPQDD171 pKa = 3.42LRR173 pKa = 11.84KK174 pKa = 10.12GQGDD178 pKa = 3.69RR179 pKa = 11.84LVLRR183 pKa = 11.84TKK185 pKa = 10.68GVLRR189 pKa = 11.84FHH191 pKa = 6.96GRR193 pKa = 11.84PKK195 pKa = 10.14TLEE198 pKa = 3.93LKK200 pKa = 10.2YY201 pKa = 11.07DD202 pKa = 3.65PVKK205 pKa = 10.08TRR207 pKa = 11.84WYY209 pKa = 7.9AHH211 pKa = 5.21QVVEE215 pKa = 4.35VPEE218 pKa = 4.41PVRR221 pKa = 11.84KK222 pKa = 9.53ARR224 pKa = 11.84PEE226 pKa = 3.42KK227 pKa = 10.39HH228 pKa = 6.06AAIDD232 pKa = 3.44LGARR236 pKa = 11.84VLVALAVEE244 pKa = 4.52GLNRR248 pKa = 11.84QLLFSARR255 pKa = 11.84EE256 pKa = 3.87VIKK259 pKa = 10.97DD260 pKa = 3.21FLYY263 pKa = 8.04WTNQIAGEE271 pKa = 3.94QSRR274 pKa = 11.84LNRR277 pKa = 11.84AGRR280 pKa = 11.84KK281 pKa = 5.74TSRR284 pKa = 11.84RR285 pKa = 11.84LKK287 pKa = 10.42RR288 pKa = 11.84LYY290 pKa = 9.27QLRR293 pKa = 11.84ARR295 pKa = 11.84RR296 pKa = 11.84LRR298 pKa = 11.84HH299 pKa = 5.83AFVALAAEE307 pKa = 4.62IVRR310 pKa = 11.84ILKK313 pKa = 10.1RR314 pKa = 11.84LRR316 pKa = 11.84VTTLFLEE323 pKa = 4.57DD324 pKa = 3.17LTGIRR329 pKa = 11.84EE330 pKa = 4.28AMDD333 pKa = 3.97FGPKK337 pKa = 9.82NLLVHH342 pKa = 6.29NFWAFRR348 pKa = 11.84MLRR351 pKa = 11.84NLIEE355 pKa = 4.26AACTRR360 pKa = 11.84TGIRR364 pKa = 11.84VVPVEE369 pKa = 4.28PRR371 pKa = 11.84GTSCRR376 pKa = 11.84CAVCGSSVRR385 pKa = 11.84RR386 pKa = 11.84PVRR389 pKa = 11.84HH390 pKa = 5.98KK391 pKa = 10.61AVCEE395 pKa = 3.58KK396 pKa = 10.6CGRR399 pKa = 11.84VWHH402 pKa = 7.0ADD404 pKa = 2.81ANAALNIILQGSSKK418 pKa = 10.18EE419 pKa = 4.16HH420 pKa = 5.99GAEE423 pKa = 3.89ATPLKK428 pKa = 10.28PLALRR433 pKa = 11.84WDD435 pKa = 3.41RR436 pKa = 11.84HH437 pKa = 4.06RR438 pKa = 11.84WVSRR442 pKa = 11.84FEE444 pKa = 4.28SAAGTPSGATSGSRR458 pKa = 11.84MAAA461 pKa = 3.18
MM1 pKa = 7.07GHH3 pKa = 6.96KK4 pKa = 9.87NLKK7 pKa = 9.57QLDD10 pKa = 4.39AIAFKK15 pKa = 10.69PGDD18 pKa = 3.4EE19 pKa = 4.27SGFRR23 pKa = 11.84LCYY26 pKa = 9.39NGSMKK31 pKa = 9.09HH32 pKa = 4.66TNVVRR37 pKa = 11.84VLPDD41 pKa = 3.1KK42 pKa = 10.6RR43 pKa = 11.84QKK45 pKa = 10.27QVLEE49 pKa = 4.47IIGDD53 pKa = 3.87RR54 pKa = 11.84CAALWNAVQYY64 pKa = 10.49RR65 pKa = 11.84CRR67 pKa = 11.84QAFFKK72 pKa = 10.78GDD74 pKa = 3.82SVPSYY79 pKa = 11.1AALCSEE85 pKa = 4.81FKK87 pKa = 10.5EE88 pKa = 4.38HH89 pKa = 6.94PAYY92 pKa = 9.76KK93 pKa = 10.27ALPAHH98 pKa = 7.2IGQEE102 pKa = 4.36VIKK105 pKa = 10.0KK106 pKa = 9.63ARR108 pKa = 11.84KK109 pKa = 8.86AWDD112 pKa = 3.36SYY114 pKa = 9.23FACLRR119 pKa = 11.84LYY121 pKa = 10.7RR122 pKa = 11.84KK123 pKa = 10.33GEE125 pKa = 4.15LKK127 pKa = 10.46EE128 pKa = 4.15PPHH131 pKa = 7.12IPRR134 pKa = 11.84YY135 pKa = 8.09WKK137 pKa = 10.1DD138 pKa = 2.87RR139 pKa = 11.84RR140 pKa = 11.84TGRR143 pKa = 11.84RR144 pKa = 11.84IFRR147 pKa = 11.84VIPVKK152 pKa = 10.88APTSYY157 pKa = 11.47SLDD160 pKa = 3.52ARR162 pKa = 11.84CLSLTLPQDD171 pKa = 3.42LRR173 pKa = 11.84KK174 pKa = 10.12GQGDD178 pKa = 3.69RR179 pKa = 11.84LVLRR183 pKa = 11.84TKK185 pKa = 10.68GVLRR189 pKa = 11.84FHH191 pKa = 6.96GRR193 pKa = 11.84PKK195 pKa = 10.14TLEE198 pKa = 3.93LKK200 pKa = 10.2YY201 pKa = 11.07DD202 pKa = 3.65PVKK205 pKa = 10.08TRR207 pKa = 11.84WYY209 pKa = 7.9AHH211 pKa = 5.21QVVEE215 pKa = 4.35VPEE218 pKa = 4.41PVRR221 pKa = 11.84KK222 pKa = 9.53ARR224 pKa = 11.84PEE226 pKa = 3.42KK227 pKa = 10.39HH228 pKa = 6.06AAIDD232 pKa = 3.44LGARR236 pKa = 11.84VLVALAVEE244 pKa = 4.52GLNRR248 pKa = 11.84QLLFSARR255 pKa = 11.84EE256 pKa = 3.87VIKK259 pKa = 10.97DD260 pKa = 3.21FLYY263 pKa = 8.04WTNQIAGEE271 pKa = 3.94QSRR274 pKa = 11.84LNRR277 pKa = 11.84AGRR280 pKa = 11.84KK281 pKa = 5.74TSRR284 pKa = 11.84RR285 pKa = 11.84LKK287 pKa = 10.42RR288 pKa = 11.84LYY290 pKa = 9.27QLRR293 pKa = 11.84ARR295 pKa = 11.84RR296 pKa = 11.84LRR298 pKa = 11.84HH299 pKa = 5.83AFVALAAEE307 pKa = 4.62IVRR310 pKa = 11.84ILKK313 pKa = 10.1RR314 pKa = 11.84LRR316 pKa = 11.84VTTLFLEE323 pKa = 4.57DD324 pKa = 3.17LTGIRR329 pKa = 11.84EE330 pKa = 4.28AMDD333 pKa = 3.97FGPKK337 pKa = 9.82NLLVHH342 pKa = 6.29NFWAFRR348 pKa = 11.84MLRR351 pKa = 11.84NLIEE355 pKa = 4.26AACTRR360 pKa = 11.84TGIRR364 pKa = 11.84VVPVEE369 pKa = 4.28PRR371 pKa = 11.84GTSCRR376 pKa = 11.84CAVCGSSVRR385 pKa = 11.84RR386 pKa = 11.84PVRR389 pKa = 11.84HH390 pKa = 5.98KK391 pKa = 10.61AVCEE395 pKa = 3.58KK396 pKa = 10.6CGRR399 pKa = 11.84VWHH402 pKa = 7.0ADD404 pKa = 2.81ANAALNIILQGSSKK418 pKa = 10.18EE419 pKa = 4.16HH420 pKa = 5.99GAEE423 pKa = 3.89ATPLKK428 pKa = 10.28PLALRR433 pKa = 11.84WDD435 pKa = 3.41RR436 pKa = 11.84HH437 pKa = 4.06RR438 pKa = 11.84WVSRR442 pKa = 11.84FEE444 pKa = 4.28SAAGTPSGATSGSRR458 pKa = 11.84MAAA461 pKa = 3.18
Molecular weight: 52.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
709530 |
30 |
2290 |
293.1 |
32.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.773 ± 0.059 | 1.063 ± 0.023 |
4.862 ± 0.031 | 8.074 ± 0.066 |
4.242 ± 0.04 | 7.569 ± 0.044 |
1.472 ± 0.02 | 7.876 ± 0.045 |
7.745 ± 0.046 | 9.773 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.629 ± 0.023 | 3.703 ± 0.028 |
4.007 ± 0.029 | 2.376 ± 0.024 |
5.179 ± 0.044 | 5.298 ± 0.033 |
4.471 ± 0.036 | 7.789 ± 0.047 |
0.786 ± 0.017 | 3.316 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
