
Ctenophore-associated circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.56
Get precalculated fractions of proteins
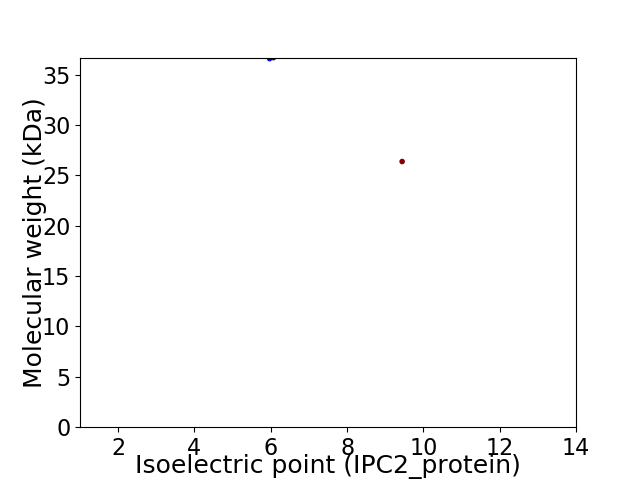
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
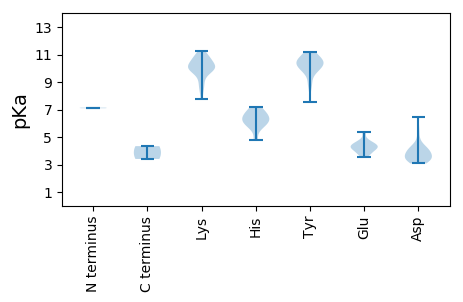
>tr|A0A141MJA6|A0A141MJA6_9VIRU Uncharacterized protein OS=Ctenophore-associated circular virus 1 OX=1778558 PE=4 SV=1
MM1 pKa = 7.14ATSEE5 pKa = 4.13TSEE8 pKa = 4.38TSGGPPPSGTFRR20 pKa = 11.84AICFTEE26 pKa = 5.28FGDD29 pKa = 4.07PPDD32 pKa = 3.6WDD34 pKa = 4.06VVQHH38 pKa = 6.44KK39 pKa = 10.04LRR41 pKa = 11.84FMAYY45 pKa = 9.97GLEE48 pKa = 4.34TCPTTQRR55 pKa = 11.84EE56 pKa = 4.58HH57 pKa = 5.77YY58 pKa = 10.25QGFAYY63 pKa = 10.02AHH65 pKa = 6.7KK66 pKa = 10.09PMRR69 pKa = 11.84WKK71 pKa = 10.12AWKK74 pKa = 10.01KK75 pKa = 9.43LFPRR79 pKa = 11.84AHH81 pKa = 6.13FEE83 pKa = 3.85KK84 pKa = 10.25MAGSFQDD91 pKa = 3.41NVVYY95 pKa = 10.48CSKK98 pKa = 10.51QSSLIKK104 pKa = 10.51FGEE107 pKa = 4.18EE108 pKa = 3.54PKK110 pKa = 10.6QGEE113 pKa = 4.39RR114 pKa = 11.84KK115 pKa = 10.14DD116 pKa = 5.07LIDD119 pKa = 4.46LKK121 pKa = 11.04RR122 pKa = 11.84KK123 pKa = 9.85LEE125 pKa = 4.27EE126 pKa = 3.65YY127 pKa = 10.45APRR130 pKa = 11.84RR131 pKa = 11.84GTTTYY136 pKa = 11.21DD137 pKa = 3.24VAEE140 pKa = 4.6DD141 pKa = 3.36EE142 pKa = 5.35AYY144 pKa = 9.76FGVVARR150 pKa = 11.84HH151 pKa = 4.75NTFASGYY158 pKa = 8.79LRR160 pKa = 11.84HH161 pKa = 6.26KK162 pKa = 9.7RR163 pKa = 11.84AKK165 pKa = 9.74ILNGDD170 pKa = 3.15RR171 pKa = 11.84TMPKK175 pKa = 8.56VYY177 pKa = 10.47VRR179 pKa = 11.84IGPPGTGKK187 pKa = 8.13TRR189 pKa = 11.84WLDD192 pKa = 3.31RR193 pKa = 11.84KK194 pKa = 9.97FGVGGYY200 pKa = 9.89VIAPDD205 pKa = 3.69NTGKK209 pKa = 9.72WFDD212 pKa = 3.85GCDD215 pKa = 3.23HH216 pKa = 7.2DD217 pKa = 6.45VILFDD222 pKa = 5.59DD223 pKa = 4.22VEE225 pKa = 4.81IDD227 pKa = 3.57QVPPLSLWKK236 pKa = 9.9RR237 pKa = 11.84LCDD240 pKa = 3.6RR241 pKa = 11.84YY242 pKa = 10.39PFEE245 pKa = 4.62VPVKK249 pKa = 10.65GGFITWKK256 pKa = 9.0PRR258 pKa = 11.84VIVFTSNHH266 pKa = 6.08AVHH269 pKa = 6.69EE270 pKa = 4.25WWKK273 pKa = 10.97DD274 pKa = 3.1ITPDD278 pKa = 3.36NQQAIARR285 pKa = 11.84RR286 pKa = 11.84IYY288 pKa = 8.89KK289 pKa = 8.55TVYY292 pKa = 10.41VDD294 pKa = 3.49GTDD297 pKa = 3.39GDD299 pKa = 4.0EE300 pKa = 4.34TQEE303 pKa = 4.0EE304 pKa = 4.47FQSDD308 pKa = 3.97EE309 pKa = 4.23DD310 pKa = 4.24SEE312 pKa = 4.46GAEE315 pKa = 4.17EE316 pKa = 4.78GRR318 pKa = 11.84AA319 pKa = 3.4
MM1 pKa = 7.14ATSEE5 pKa = 4.13TSEE8 pKa = 4.38TSGGPPPSGTFRR20 pKa = 11.84AICFTEE26 pKa = 5.28FGDD29 pKa = 4.07PPDD32 pKa = 3.6WDD34 pKa = 4.06VVQHH38 pKa = 6.44KK39 pKa = 10.04LRR41 pKa = 11.84FMAYY45 pKa = 9.97GLEE48 pKa = 4.34TCPTTQRR55 pKa = 11.84EE56 pKa = 4.58HH57 pKa = 5.77YY58 pKa = 10.25QGFAYY63 pKa = 10.02AHH65 pKa = 6.7KK66 pKa = 10.09PMRR69 pKa = 11.84WKK71 pKa = 10.12AWKK74 pKa = 10.01KK75 pKa = 9.43LFPRR79 pKa = 11.84AHH81 pKa = 6.13FEE83 pKa = 3.85KK84 pKa = 10.25MAGSFQDD91 pKa = 3.41NVVYY95 pKa = 10.48CSKK98 pKa = 10.51QSSLIKK104 pKa = 10.51FGEE107 pKa = 4.18EE108 pKa = 3.54PKK110 pKa = 10.6QGEE113 pKa = 4.39RR114 pKa = 11.84KK115 pKa = 10.14DD116 pKa = 5.07LIDD119 pKa = 4.46LKK121 pKa = 11.04RR122 pKa = 11.84KK123 pKa = 9.85LEE125 pKa = 4.27EE126 pKa = 3.65YY127 pKa = 10.45APRR130 pKa = 11.84RR131 pKa = 11.84GTTTYY136 pKa = 11.21DD137 pKa = 3.24VAEE140 pKa = 4.6DD141 pKa = 3.36EE142 pKa = 5.35AYY144 pKa = 9.76FGVVARR150 pKa = 11.84HH151 pKa = 4.75NTFASGYY158 pKa = 8.79LRR160 pKa = 11.84HH161 pKa = 6.26KK162 pKa = 9.7RR163 pKa = 11.84AKK165 pKa = 9.74ILNGDD170 pKa = 3.15RR171 pKa = 11.84TMPKK175 pKa = 8.56VYY177 pKa = 10.47VRR179 pKa = 11.84IGPPGTGKK187 pKa = 8.13TRR189 pKa = 11.84WLDD192 pKa = 3.31RR193 pKa = 11.84KK194 pKa = 9.97FGVGGYY200 pKa = 9.89VIAPDD205 pKa = 3.69NTGKK209 pKa = 9.72WFDD212 pKa = 3.85GCDD215 pKa = 3.23HH216 pKa = 7.2DD217 pKa = 6.45VILFDD222 pKa = 5.59DD223 pKa = 4.22VEE225 pKa = 4.81IDD227 pKa = 3.57QVPPLSLWKK236 pKa = 9.9RR237 pKa = 11.84LCDD240 pKa = 3.6RR241 pKa = 11.84YY242 pKa = 10.39PFEE245 pKa = 4.62VPVKK249 pKa = 10.65GGFITWKK256 pKa = 9.0PRR258 pKa = 11.84VIVFTSNHH266 pKa = 6.08AVHH269 pKa = 6.69EE270 pKa = 4.25WWKK273 pKa = 10.97DD274 pKa = 3.1ITPDD278 pKa = 3.36NQQAIARR285 pKa = 11.84RR286 pKa = 11.84IYY288 pKa = 8.89KK289 pKa = 8.55TVYY292 pKa = 10.41VDD294 pKa = 3.49GTDD297 pKa = 3.39GDD299 pKa = 4.0EE300 pKa = 4.34TQEE303 pKa = 4.0EE304 pKa = 4.47FQSDD308 pKa = 3.97EE309 pKa = 4.23DD310 pKa = 4.24SEE312 pKa = 4.46GAEE315 pKa = 4.17EE316 pKa = 4.78GRR318 pKa = 11.84AA319 pKa = 3.4
Molecular weight: 36.59 kDa
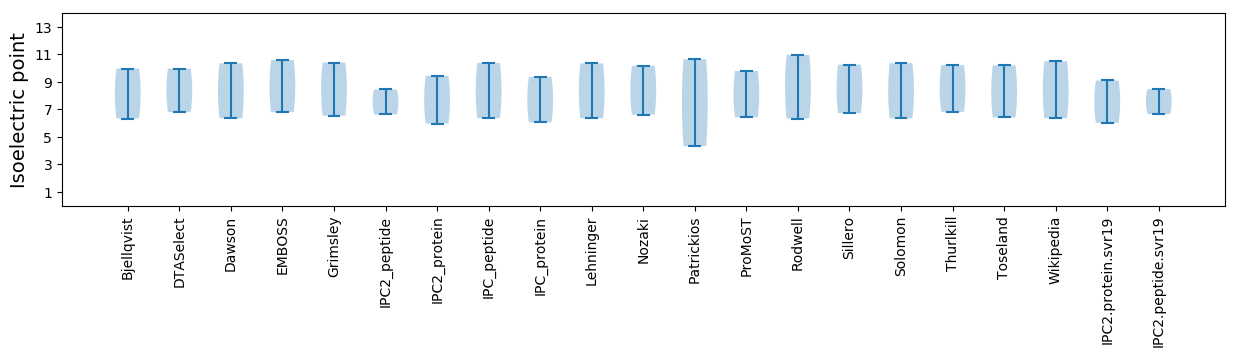
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A141MJA6|A0A141MJA6_9VIRU Uncharacterized protein OS=Ctenophore-associated circular virus 1 OX=1778558 PE=4 SV=1
MM1 pKa = 7.09KK2 pKa = 10.09RR3 pKa = 11.84KK4 pKa = 9.62RR5 pKa = 11.84SSSQTRR11 pKa = 11.84TVKK14 pKa = 9.9VRR16 pKa = 11.84KK17 pKa = 9.17KK18 pKa = 10.1AAPKK22 pKa = 10.13TMTAQRR28 pKa = 11.84RR29 pKa = 11.84LIRR32 pKa = 11.84RR33 pKa = 11.84MILKK37 pKa = 7.76TTEE40 pKa = 3.95TKK42 pKa = 10.87YY43 pKa = 9.72KK44 pKa = 10.2TIALDD49 pKa = 3.55KK50 pKa = 11.27EE51 pKa = 4.42EE52 pKa = 4.59IYY54 pKa = 10.9HH55 pKa = 5.76NKK57 pKa = 10.21LYY59 pKa = 10.97NFSKK63 pKa = 10.82NLLEE67 pKa = 4.25VFPSQGTGDD76 pKa = 3.84GARR79 pKa = 11.84EE80 pKa = 3.74GDD82 pKa = 3.96EE83 pKa = 4.05IVTTGMKK90 pKa = 10.09VRR92 pKa = 11.84MILGQKK98 pKa = 9.07SDD100 pKa = 4.11RR101 pKa = 11.84PNVSYY106 pKa = 10.62KK107 pKa = 10.28VWVVHH112 pKa = 6.3YY113 pKa = 10.56DD114 pKa = 3.44DD115 pKa = 4.53STSGNAITYY124 pKa = 10.61GNFFHH129 pKa = 6.92NVTGNGMLDD138 pKa = 3.08AVQNKK143 pKa = 8.65RR144 pKa = 11.84FKK146 pKa = 10.43VLKK149 pKa = 10.55AFTIKK154 pKa = 10.8SKK156 pKa = 9.6GTTIEE161 pKa = 3.9AGEE164 pKa = 4.08TSKK167 pKa = 11.26EE168 pKa = 3.68FVRR171 pKa = 11.84PVKK174 pKa = 10.54FWIPFKK180 pKa = 10.9KK181 pKa = 10.03KK182 pKa = 10.52LKK184 pKa = 9.91FVSDD188 pKa = 4.3ASNKK192 pKa = 9.88ASNILPQMNVIVLPYY207 pKa = 10.05DD208 pKa = 3.47AYY210 pKa = 10.35GTLGTDD216 pKa = 4.01NIAYY220 pKa = 7.58MQGSCTLYY228 pKa = 11.06YY229 pKa = 10.38KK230 pKa = 10.8DD231 pKa = 3.73PP232 pKa = 4.34
MM1 pKa = 7.09KK2 pKa = 10.09RR3 pKa = 11.84KK4 pKa = 9.62RR5 pKa = 11.84SSSQTRR11 pKa = 11.84TVKK14 pKa = 9.9VRR16 pKa = 11.84KK17 pKa = 9.17KK18 pKa = 10.1AAPKK22 pKa = 10.13TMTAQRR28 pKa = 11.84RR29 pKa = 11.84LIRR32 pKa = 11.84RR33 pKa = 11.84MILKK37 pKa = 7.76TTEE40 pKa = 3.95TKK42 pKa = 10.87YY43 pKa = 9.72KK44 pKa = 10.2TIALDD49 pKa = 3.55KK50 pKa = 11.27EE51 pKa = 4.42EE52 pKa = 4.59IYY54 pKa = 10.9HH55 pKa = 5.76NKK57 pKa = 10.21LYY59 pKa = 10.97NFSKK63 pKa = 10.82NLLEE67 pKa = 4.25VFPSQGTGDD76 pKa = 3.84GARR79 pKa = 11.84EE80 pKa = 3.74GDD82 pKa = 3.96EE83 pKa = 4.05IVTTGMKK90 pKa = 10.09VRR92 pKa = 11.84MILGQKK98 pKa = 9.07SDD100 pKa = 4.11RR101 pKa = 11.84PNVSYY106 pKa = 10.62KK107 pKa = 10.28VWVVHH112 pKa = 6.3YY113 pKa = 10.56DD114 pKa = 3.44DD115 pKa = 4.53STSGNAITYY124 pKa = 10.61GNFFHH129 pKa = 6.92NVTGNGMLDD138 pKa = 3.08AVQNKK143 pKa = 8.65RR144 pKa = 11.84FKK146 pKa = 10.43VLKK149 pKa = 10.55AFTIKK154 pKa = 10.8SKK156 pKa = 9.6GTTIEE161 pKa = 3.9AGEE164 pKa = 4.08TSKK167 pKa = 11.26EE168 pKa = 3.68FVRR171 pKa = 11.84PVKK174 pKa = 10.54FWIPFKK180 pKa = 10.9KK181 pKa = 10.03KK182 pKa = 10.52LKK184 pKa = 9.91FVSDD188 pKa = 4.3ASNKK192 pKa = 9.88ASNILPQMNVIVLPYY207 pKa = 10.05DD208 pKa = 3.47AYY210 pKa = 10.35GTLGTDD216 pKa = 4.01NIAYY220 pKa = 7.58MQGSCTLYY228 pKa = 11.06YY229 pKa = 10.38KK230 pKa = 10.8DD231 pKa = 3.73PP232 pKa = 4.34
Molecular weight: 26.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
551 |
232 |
319 |
275.5 |
31.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.989 ± 0.237 | 1.089 ± 0.404 |
6.534 ± 1.101 | 5.808 ± 1.185 |
5.082 ± 0.474 | 7.623 ± 0.711 |
2.178 ± 0.544 | 4.719 ± 0.544 |
9.256 ± 1.729 | 5.082 ± 0.585 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.359 ± 0.669 | 3.448 ± 1.324 |
5.082 ± 1.004 | 3.267 ± 0.153 |
6.352 ± 0.46 | 5.082 ± 0.85 |
7.804 ± 0.767 | 6.897 ± 0.265 |
1.996 ± 0.697 | 4.356 ± 0.237 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
