
Sanxia sobemo-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
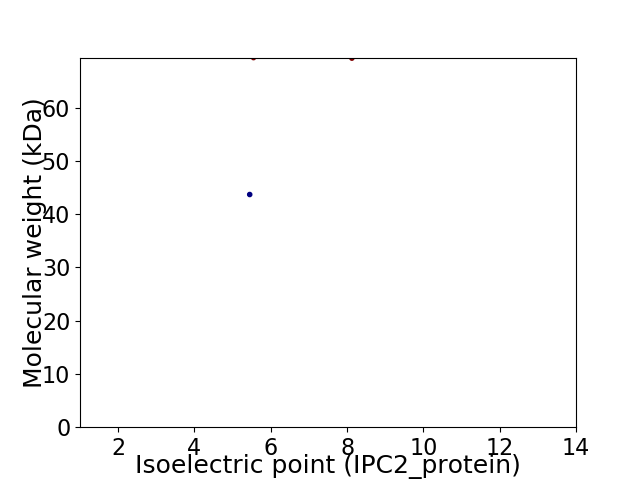
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KER0|A0A1L3KER0_9VIRU RNA-directed RNA polymerase OS=Sanxia sobemo-like virus 3 OX=1923382 PE=4 SV=1
MM1 pKa = 7.15NAVEE5 pKa = 4.54VFSSLRR11 pKa = 11.84WEE13 pKa = 4.36IPDD16 pKa = 4.79DD17 pKa = 3.88FMTYY21 pKa = 7.92SHH23 pKa = 7.71FLRR26 pKa = 11.84VLDD29 pKa = 4.29KK30 pKa = 11.4LDD32 pKa = 3.58MASSPGYY39 pKa = 9.31PYY41 pKa = 9.92MRR43 pKa = 11.84RR44 pKa = 11.84APVNKK49 pKa = 9.19ILFRR53 pKa = 11.84VDD55 pKa = 2.99EE56 pKa = 4.75EE57 pKa = 4.8GAMNPSSVDD66 pKa = 3.68YY67 pKa = 10.48IWQTVSQRR75 pKa = 11.84IAEE78 pKa = 4.5FGDD81 pKa = 3.6ADD83 pKa = 3.71PVRR86 pKa = 11.84FFIKK90 pKa = 10.38QEE92 pKa = 3.58PHH94 pKa = 6.15KK95 pKa = 10.65LKK97 pKa = 10.88KK98 pKa = 10.64LVDD101 pKa = 3.42GRR103 pKa = 11.84NRR105 pKa = 11.84LISSVSVVDD114 pKa = 4.52QIIDD118 pKa = 3.35HH119 pKa = 6.25MLFDD123 pKa = 4.43SMNEE127 pKa = 3.77NMVDD131 pKa = 2.95NWNYY135 pKa = 10.69NPIKK139 pKa = 10.44IGWSPVKK146 pKa = 10.5GGWKK150 pKa = 10.15AFPTGKK156 pKa = 10.16RR157 pKa = 11.84IAIDD161 pKa = 3.22KK162 pKa = 9.97SAWDD166 pKa = 3.48WTVQMWMLEE175 pKa = 3.97LCLTIRR181 pKa = 11.84LALCRR186 pKa = 11.84TDD188 pKa = 4.04GPNRR192 pKa = 11.84DD193 pKa = 3.78LWLKK197 pKa = 9.6IAIYY201 pKa = 9.86RR202 pKa = 11.84YY203 pKa = 9.53KK204 pKa = 10.47QLFLHH209 pKa = 6.59PLFITSGGLLLRR221 pKa = 11.84QQQEE225 pKa = 4.63GVQKK229 pKa = 10.3SGCVNTLADD238 pKa = 3.89NSLMQWILHH247 pKa = 5.71ARR249 pKa = 11.84VCLEE253 pKa = 3.9HH254 pKa = 7.39GVDD257 pKa = 4.7IEE259 pKa = 4.44DD260 pKa = 4.11DD261 pKa = 4.07EE262 pKa = 4.53MWVMGDD268 pKa = 3.92DD269 pKa = 4.19TSQSAVPDD277 pKa = 4.64GYY279 pKa = 11.16EE280 pKa = 3.58DD281 pKa = 3.38WLRR284 pKa = 11.84EE285 pKa = 3.77YY286 pKa = 10.75CIVKK290 pKa = 9.29EE291 pKa = 4.1CQPARR296 pKa = 11.84EE297 pKa = 4.05FCGFRR302 pKa = 11.84FDD304 pKa = 4.98RR305 pKa = 11.84IHH307 pKa = 7.4VEE309 pKa = 3.5PLYY312 pKa = 10.6KK313 pKa = 10.4GKK315 pKa = 9.54HH316 pKa = 5.09AFNLLHH322 pKa = 7.35LDD324 pKa = 3.61EE325 pKa = 5.45KK326 pKa = 11.08NAAEE330 pKa = 4.28VATSYY335 pKa = 11.69ALLYY339 pKa = 10.06HH340 pKa = 7.14RR341 pKa = 11.84SRR343 pKa = 11.84HH344 pKa = 5.5RR345 pKa = 11.84DD346 pKa = 2.42WMRR349 pKa = 11.84NLFEE353 pKa = 5.11SMSLNIFPLWYY364 pKa = 10.16YY365 pKa = 10.65DD366 pKa = 3.98AIYY369 pKa = 10.87DD370 pKa = 4.1GVEE373 pKa = 3.52
MM1 pKa = 7.15NAVEE5 pKa = 4.54VFSSLRR11 pKa = 11.84WEE13 pKa = 4.36IPDD16 pKa = 4.79DD17 pKa = 3.88FMTYY21 pKa = 7.92SHH23 pKa = 7.71FLRR26 pKa = 11.84VLDD29 pKa = 4.29KK30 pKa = 11.4LDD32 pKa = 3.58MASSPGYY39 pKa = 9.31PYY41 pKa = 9.92MRR43 pKa = 11.84RR44 pKa = 11.84APVNKK49 pKa = 9.19ILFRR53 pKa = 11.84VDD55 pKa = 2.99EE56 pKa = 4.75EE57 pKa = 4.8GAMNPSSVDD66 pKa = 3.68YY67 pKa = 10.48IWQTVSQRR75 pKa = 11.84IAEE78 pKa = 4.5FGDD81 pKa = 3.6ADD83 pKa = 3.71PVRR86 pKa = 11.84FFIKK90 pKa = 10.38QEE92 pKa = 3.58PHH94 pKa = 6.15KK95 pKa = 10.65LKK97 pKa = 10.88KK98 pKa = 10.64LVDD101 pKa = 3.42GRR103 pKa = 11.84NRR105 pKa = 11.84LISSVSVVDD114 pKa = 4.52QIIDD118 pKa = 3.35HH119 pKa = 6.25MLFDD123 pKa = 4.43SMNEE127 pKa = 3.77NMVDD131 pKa = 2.95NWNYY135 pKa = 10.69NPIKK139 pKa = 10.44IGWSPVKK146 pKa = 10.5GGWKK150 pKa = 10.15AFPTGKK156 pKa = 10.16RR157 pKa = 11.84IAIDD161 pKa = 3.22KK162 pKa = 9.97SAWDD166 pKa = 3.48WTVQMWMLEE175 pKa = 3.97LCLTIRR181 pKa = 11.84LALCRR186 pKa = 11.84TDD188 pKa = 4.04GPNRR192 pKa = 11.84DD193 pKa = 3.78LWLKK197 pKa = 9.6IAIYY201 pKa = 9.86RR202 pKa = 11.84YY203 pKa = 9.53KK204 pKa = 10.47QLFLHH209 pKa = 6.59PLFITSGGLLLRR221 pKa = 11.84QQQEE225 pKa = 4.63GVQKK229 pKa = 10.3SGCVNTLADD238 pKa = 3.89NSLMQWILHH247 pKa = 5.71ARR249 pKa = 11.84VCLEE253 pKa = 3.9HH254 pKa = 7.39GVDD257 pKa = 4.7IEE259 pKa = 4.44DD260 pKa = 4.11DD261 pKa = 4.07EE262 pKa = 4.53MWVMGDD268 pKa = 3.92DD269 pKa = 4.19TSQSAVPDD277 pKa = 4.64GYY279 pKa = 11.16EE280 pKa = 3.58DD281 pKa = 3.38WLRR284 pKa = 11.84EE285 pKa = 3.77YY286 pKa = 10.75CIVKK290 pKa = 9.29EE291 pKa = 4.1CQPARR296 pKa = 11.84EE297 pKa = 4.05FCGFRR302 pKa = 11.84FDD304 pKa = 4.98RR305 pKa = 11.84IHH307 pKa = 7.4VEE309 pKa = 3.5PLYY312 pKa = 10.6KK313 pKa = 10.4GKK315 pKa = 9.54HH316 pKa = 5.09AFNLLHH322 pKa = 7.35LDD324 pKa = 3.61EE325 pKa = 5.45KK326 pKa = 11.08NAAEE330 pKa = 4.28VATSYY335 pKa = 11.69ALLYY339 pKa = 10.06HH340 pKa = 7.14RR341 pKa = 11.84SRR343 pKa = 11.84HH344 pKa = 5.5RR345 pKa = 11.84DD346 pKa = 2.42WMRR349 pKa = 11.84NLFEE353 pKa = 5.11SMSLNIFPLWYY364 pKa = 10.16YY365 pKa = 10.65DD366 pKa = 3.98AIYY369 pKa = 10.87DD370 pKa = 4.1GVEE373 pKa = 3.52
Molecular weight: 43.67 kDa
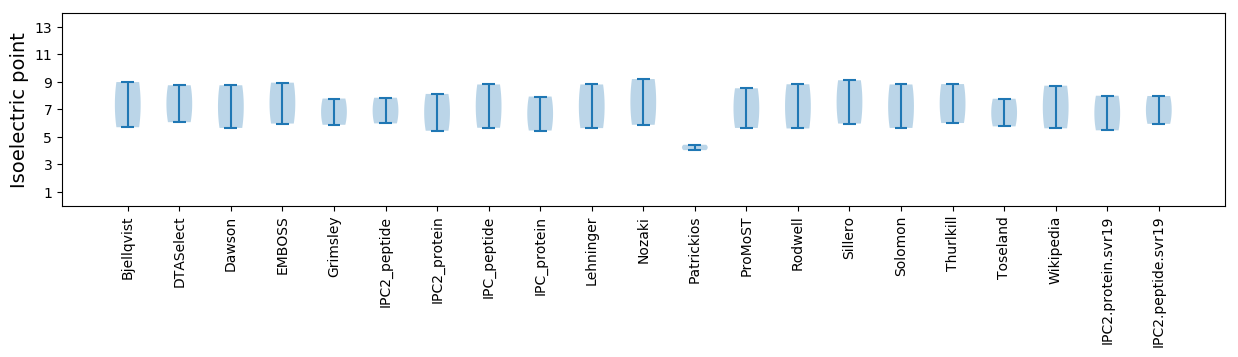
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KER0|A0A1L3KER0_9VIRU RNA-directed RNA polymerase OS=Sanxia sobemo-like virus 3 OX=1923382 PE=4 SV=1
MM1 pKa = 7.26AQNEE5 pKa = 4.41GNRR8 pKa = 11.84TNFLTEE14 pKa = 3.36ISDD17 pKa = 3.53TYY19 pKa = 10.81TRR21 pKa = 11.84ITRR24 pKa = 11.84AASLMVRR31 pKa = 11.84IGIYY35 pKa = 10.05YY36 pKa = 7.67ATAVSFCLLYY46 pKa = 10.86RR47 pKa = 11.84LTRR50 pKa = 11.84WVLSQPVTWTLNAAEE65 pKa = 4.25FVEE68 pKa = 4.64HH69 pKa = 6.11VFYY72 pKa = 10.95GVGTWALWPLDD83 pKa = 3.57KK84 pKa = 11.26GADD87 pKa = 3.14IVVSFLSGPKK97 pKa = 9.59PEE99 pKa = 4.11PTVTEE104 pKa = 4.57RR105 pKa = 11.84IIEE108 pKa = 4.21EE109 pKa = 4.15VNEE112 pKa = 4.02KK113 pKa = 11.0AEE115 pKa = 3.99YY116 pKa = 10.05LYY118 pKa = 11.17SFSAYY123 pKa = 10.2LPSLCYY129 pKa = 9.03ITMVVLIVLLLWVLSRR145 pKa = 11.84RR146 pKa = 11.84YY147 pKa = 10.33LRR149 pKa = 11.84TTLMRR154 pKa = 11.84MRR156 pKa = 11.84GVYY159 pKa = 9.71IGEE162 pKa = 4.14SMRR165 pKa = 11.84NGSKK169 pKa = 10.5FMPAQIPSGQVAVMSAGMLVDD190 pKa = 3.02NHH192 pKa = 6.55IGYY195 pKa = 9.8GLRR198 pKa = 11.84VGNVLVTPYY207 pKa = 10.56HH208 pKa = 6.5VIRR211 pKa = 11.84DD212 pKa = 3.7IVNPILSYY220 pKa = 10.61GGKK223 pKa = 9.74KK224 pKa = 9.17ICVNVAHH231 pKa = 6.31VVASKK236 pKa = 10.15CIPDD240 pKa = 3.59VAYY243 pKa = 11.24LMLDD247 pKa = 3.19EE248 pKa = 4.6TQWTRR253 pKa = 11.84LGVPKK258 pKa = 10.66VKK260 pKa = 10.55VVTKK264 pKa = 5.83TTNVATSVTCVGLSGQSVGLLRR286 pKa = 11.84KK287 pKa = 9.56SSRR290 pKa = 11.84IGMMIYY296 pKa = 10.12SGSTIPGMSGAGYY309 pKa = 9.42FVNNQCHH316 pKa = 5.38GMHH319 pKa = 6.71NGVIMHH325 pKa = 7.03DD326 pKa = 3.54NVGVSMSCIYY336 pKa = 11.12GEE338 pKa = 4.1LKK340 pKa = 10.17MIFRR344 pKa = 11.84GEE346 pKa = 4.05SSHH349 pKa = 7.42DD350 pKa = 3.56FGEE353 pKa = 4.41EE354 pKa = 4.14VVKK357 pKa = 10.8KK358 pKa = 10.25SAGVRR363 pKa = 11.84IFKK366 pKa = 8.78TWSDD370 pKa = 3.49EE371 pKa = 4.18DD372 pKa = 3.85LVKK375 pKa = 10.6QFHH378 pKa = 6.24EE379 pKa = 4.49MWQEE383 pKa = 3.49PDD385 pKa = 3.41FEE387 pKa = 4.71EE388 pKa = 5.87DD389 pKa = 3.88ISGAWGMRR397 pKa = 11.84LGEE400 pKa = 4.07EE401 pKa = 4.22SADD404 pKa = 4.17KK405 pKa = 10.46IIKK408 pKa = 8.65LTGQSPDD415 pKa = 3.41QPEE418 pKa = 3.92MVYY421 pKa = 8.91RR422 pKa = 11.84TTVEE426 pKa = 4.32CKK428 pKa = 10.19SEE430 pKa = 4.03AEE432 pKa = 3.8LRR434 pKa = 11.84EE435 pKa = 4.06RR436 pKa = 11.84IIKK439 pKa = 10.22LEE441 pKa = 3.93QVVEE445 pKa = 3.82ALKK448 pKa = 10.39IEE450 pKa = 4.43KK451 pKa = 9.92CDD453 pKa = 3.16QCSAMFVGVTLAQHH467 pKa = 6.78KK468 pKa = 8.88KK469 pKa = 6.86QHH471 pKa = 6.15SKK473 pKa = 10.58HH474 pKa = 5.85PCISCTNVFGSKK486 pKa = 9.95KK487 pKa = 10.22DD488 pKa = 3.83LEE490 pKa = 4.1QHH492 pKa = 6.17VIKK495 pKa = 9.34THH497 pKa = 4.62VKK499 pKa = 9.99YY500 pKa = 10.54PCDD503 pKa = 3.29HH504 pKa = 6.92CGVVCRR510 pKa = 11.84TEE512 pKa = 4.58VKK514 pKa = 10.36LKK516 pKa = 9.42NHH518 pKa = 6.7RR519 pKa = 11.84EE520 pKa = 4.02SCKK523 pKa = 10.34VSGGGIPMKK532 pKa = 10.36NLKK535 pKa = 10.36GEE537 pKa = 4.12SAFAMDD543 pKa = 3.75HH544 pKa = 5.91KK545 pKa = 10.81KK546 pKa = 9.96IVKK549 pKa = 8.93TDD551 pKa = 3.54PFLGQQRR558 pKa = 11.84TSRR561 pKa = 11.84KK562 pKa = 8.64KK563 pKa = 9.54NGRR566 pKa = 11.84SSEE569 pKa = 4.07RR570 pKa = 11.84TSPAKK575 pKa = 10.07VANHH579 pKa = 6.84RR580 pKa = 11.84SPSQEE585 pKa = 4.14EE586 pKa = 4.08ILCQILQSQKK596 pKa = 10.82NMQQNFEE603 pKa = 4.34KK604 pKa = 10.28FLQVMAGPKK613 pKa = 9.59PATTQNN619 pKa = 3.25
MM1 pKa = 7.26AQNEE5 pKa = 4.41GNRR8 pKa = 11.84TNFLTEE14 pKa = 3.36ISDD17 pKa = 3.53TYY19 pKa = 10.81TRR21 pKa = 11.84ITRR24 pKa = 11.84AASLMVRR31 pKa = 11.84IGIYY35 pKa = 10.05YY36 pKa = 7.67ATAVSFCLLYY46 pKa = 10.86RR47 pKa = 11.84LTRR50 pKa = 11.84WVLSQPVTWTLNAAEE65 pKa = 4.25FVEE68 pKa = 4.64HH69 pKa = 6.11VFYY72 pKa = 10.95GVGTWALWPLDD83 pKa = 3.57KK84 pKa = 11.26GADD87 pKa = 3.14IVVSFLSGPKK97 pKa = 9.59PEE99 pKa = 4.11PTVTEE104 pKa = 4.57RR105 pKa = 11.84IIEE108 pKa = 4.21EE109 pKa = 4.15VNEE112 pKa = 4.02KK113 pKa = 11.0AEE115 pKa = 3.99YY116 pKa = 10.05LYY118 pKa = 11.17SFSAYY123 pKa = 10.2LPSLCYY129 pKa = 9.03ITMVVLIVLLLWVLSRR145 pKa = 11.84RR146 pKa = 11.84YY147 pKa = 10.33LRR149 pKa = 11.84TTLMRR154 pKa = 11.84MRR156 pKa = 11.84GVYY159 pKa = 9.71IGEE162 pKa = 4.14SMRR165 pKa = 11.84NGSKK169 pKa = 10.5FMPAQIPSGQVAVMSAGMLVDD190 pKa = 3.02NHH192 pKa = 6.55IGYY195 pKa = 9.8GLRR198 pKa = 11.84VGNVLVTPYY207 pKa = 10.56HH208 pKa = 6.5VIRR211 pKa = 11.84DD212 pKa = 3.7IVNPILSYY220 pKa = 10.61GGKK223 pKa = 9.74KK224 pKa = 9.17ICVNVAHH231 pKa = 6.31VVASKK236 pKa = 10.15CIPDD240 pKa = 3.59VAYY243 pKa = 11.24LMLDD247 pKa = 3.19EE248 pKa = 4.6TQWTRR253 pKa = 11.84LGVPKK258 pKa = 10.66VKK260 pKa = 10.55VVTKK264 pKa = 5.83TTNVATSVTCVGLSGQSVGLLRR286 pKa = 11.84KK287 pKa = 9.56SSRR290 pKa = 11.84IGMMIYY296 pKa = 10.12SGSTIPGMSGAGYY309 pKa = 9.42FVNNQCHH316 pKa = 5.38GMHH319 pKa = 6.71NGVIMHH325 pKa = 7.03DD326 pKa = 3.54NVGVSMSCIYY336 pKa = 11.12GEE338 pKa = 4.1LKK340 pKa = 10.17MIFRR344 pKa = 11.84GEE346 pKa = 4.05SSHH349 pKa = 7.42DD350 pKa = 3.56FGEE353 pKa = 4.41EE354 pKa = 4.14VVKK357 pKa = 10.8KK358 pKa = 10.25SAGVRR363 pKa = 11.84IFKK366 pKa = 8.78TWSDD370 pKa = 3.49EE371 pKa = 4.18DD372 pKa = 3.85LVKK375 pKa = 10.6QFHH378 pKa = 6.24EE379 pKa = 4.49MWQEE383 pKa = 3.49PDD385 pKa = 3.41FEE387 pKa = 4.71EE388 pKa = 5.87DD389 pKa = 3.88ISGAWGMRR397 pKa = 11.84LGEE400 pKa = 4.07EE401 pKa = 4.22SADD404 pKa = 4.17KK405 pKa = 10.46IIKK408 pKa = 8.65LTGQSPDD415 pKa = 3.41QPEE418 pKa = 3.92MVYY421 pKa = 8.91RR422 pKa = 11.84TTVEE426 pKa = 4.32CKK428 pKa = 10.19SEE430 pKa = 4.03AEE432 pKa = 3.8LRR434 pKa = 11.84EE435 pKa = 4.06RR436 pKa = 11.84IIKK439 pKa = 10.22LEE441 pKa = 3.93QVVEE445 pKa = 3.82ALKK448 pKa = 10.39IEE450 pKa = 4.43KK451 pKa = 9.92CDD453 pKa = 3.16QCSAMFVGVTLAQHH467 pKa = 6.78KK468 pKa = 8.88KK469 pKa = 6.86QHH471 pKa = 6.15SKK473 pKa = 10.58HH474 pKa = 5.85PCISCTNVFGSKK486 pKa = 9.95KK487 pKa = 10.22DD488 pKa = 3.83LEE490 pKa = 4.1QHH492 pKa = 6.17VIKK495 pKa = 9.34THH497 pKa = 4.62VKK499 pKa = 9.99YY500 pKa = 10.54PCDD503 pKa = 3.29HH504 pKa = 6.92CGVVCRR510 pKa = 11.84TEE512 pKa = 4.58VKK514 pKa = 10.36LKK516 pKa = 9.42NHH518 pKa = 6.7RR519 pKa = 11.84EE520 pKa = 4.02SCKK523 pKa = 10.34VSGGGIPMKK532 pKa = 10.36NLKK535 pKa = 10.36GEE537 pKa = 4.12SAFAMDD543 pKa = 3.75HH544 pKa = 5.91KK545 pKa = 10.81KK546 pKa = 9.96IVKK549 pKa = 8.93TDD551 pKa = 3.54PFLGQQRR558 pKa = 11.84TSRR561 pKa = 11.84KK562 pKa = 8.64KK563 pKa = 9.54NGRR566 pKa = 11.84SSEE569 pKa = 4.07RR570 pKa = 11.84TSPAKK575 pKa = 10.07VANHH579 pKa = 6.84RR580 pKa = 11.84SPSQEE585 pKa = 4.14EE586 pKa = 4.08ILCQILQSQKK596 pKa = 10.82NMQQNFEE603 pKa = 4.34KK604 pKa = 10.28FLQVMAGPKK613 pKa = 9.59PATTQNN619 pKa = 3.25
Molecular weight: 69.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
992 |
373 |
619 |
496.0 |
56.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.343 ± 0.176 | 2.419 ± 0.333 |
5.04 ± 1.843 | 6.048 ± 0.257 |
3.629 ± 0.57 | 6.552 ± 0.896 |
2.923 ± 0.016 | 5.847 ± 0.032 |
6.149 ± 0.813 | 8.165 ± 0.912 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.032 ± 0.007 | 3.931 ± 0.22 |
4.032 ± 0.158 | 3.931 ± 0.274 |
5.544 ± 0.546 | 7.157 ± 0.608 |
4.738 ± 1.263 | 8.669 ± 1.208 |
2.319 ± 0.881 | 3.528 ± 0.303 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
