
Flavobacterium sediminis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
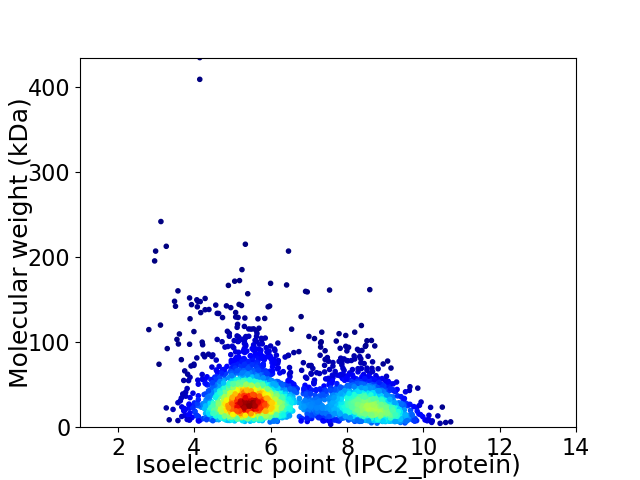
Virtual 2D-PAGE plot for 2952 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
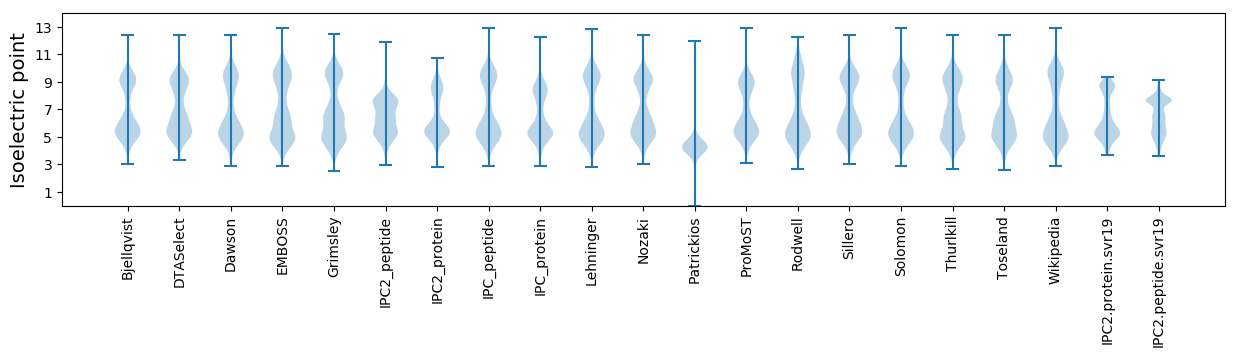
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U8QW20|A0A2U8QW20_9FLAO DNA mismatch repair protein MutL OS=Flavobacterium sediminis OX=2201181 GN=mutL PE=3 SV=1
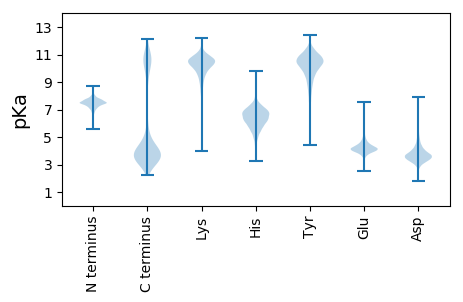
MM1 pKa = 7.65KK2 pKa = 10.51KK3 pKa = 10.51LINNNFRR10 pKa = 11.84FALPLFSLLALASCDD25 pKa = 4.35DD26 pKa = 3.95NMDD29 pKa = 3.7EE30 pKa = 4.66PGYY33 pKa = 9.44PALADD38 pKa = 3.72KK39 pKa = 10.74PVVTLTYY46 pKa = 10.98DD47 pKa = 3.24NLNILEE53 pKa = 5.09LDD55 pKa = 4.07DD56 pKa = 5.16EE57 pKa = 4.82STTGVNEE64 pKa = 4.35NIATFVLTADD74 pKa = 4.85RR75 pKa = 11.84IYY77 pKa = 11.02KK78 pKa = 10.31EE79 pKa = 4.32DD80 pKa = 3.37MKK82 pKa = 11.2FSIKK86 pKa = 10.44LNAAEE91 pKa = 4.28STADD95 pKa = 3.53AKK97 pKa = 11.28DD98 pKa = 3.62FTISLSEE105 pKa = 4.04ADD107 pKa = 4.69LDD109 pKa = 4.23YY110 pKa = 9.94GTDD113 pKa = 3.47GAFVATIPANTLSTTFTVAANFDD136 pKa = 3.95IYY138 pKa = 10.79PEE140 pKa = 4.15STEE143 pKa = 4.19NLVFDD148 pKa = 4.99IKK150 pKa = 11.07PMADD154 pKa = 3.4LNGTIAGSSKK164 pKa = 10.39KK165 pKa = 10.51VEE167 pKa = 3.97LTIGNSTSDD176 pKa = 3.97EE177 pKa = 4.15LVLILDD183 pKa = 3.83WNGDD187 pKa = 3.43STYY190 pKa = 11.49TNITDD195 pKa = 3.94PNDD198 pKa = 3.91PYY200 pKa = 11.41EE201 pKa = 4.44STLADD206 pKa = 3.45YY207 pKa = 11.32DD208 pKa = 4.13FDD210 pKa = 5.43LEE212 pKa = 4.65VYY214 pKa = 10.54NSGFSLVDD222 pKa = 3.5YY223 pKa = 10.07SWSDD227 pKa = 3.13SPEE230 pKa = 3.79QVNFATSNADD240 pKa = 2.88GLYY243 pKa = 9.95FVIAGFYY250 pKa = 8.07STYY253 pKa = 10.57GAVPMLPINFDD264 pKa = 3.45VTLTIAKK271 pKa = 9.43PGVFVYY277 pKa = 10.27DD278 pKa = 3.81YY279 pKa = 11.44DD280 pKa = 4.31MSGIWNSTDD289 pKa = 3.33GGDD292 pKa = 4.79DD293 pKa = 3.49EE294 pKa = 6.28GNVDD298 pKa = 4.06AYY300 pKa = 10.03QVPVYY305 pKa = 9.82FVKK308 pKa = 8.6TTTNGVATYY317 pKa = 9.73EE318 pKa = 4.29VYY320 pKa = 11.04NLADD324 pKa = 3.91DD325 pKa = 4.06TLLASGKK332 pKa = 10.38SSTDD336 pKa = 2.94LKK338 pKa = 11.37VYY340 pKa = 10.15FNSFKK345 pKa = 10.5RR346 pKa = 11.84VKK348 pKa = 10.42
MM1 pKa = 7.65KK2 pKa = 10.51KK3 pKa = 10.51LINNNFRR10 pKa = 11.84FALPLFSLLALASCDD25 pKa = 4.35DD26 pKa = 3.95NMDD29 pKa = 3.7EE30 pKa = 4.66PGYY33 pKa = 9.44PALADD38 pKa = 3.72KK39 pKa = 10.74PVVTLTYY46 pKa = 10.98DD47 pKa = 3.24NLNILEE53 pKa = 5.09LDD55 pKa = 4.07DD56 pKa = 5.16EE57 pKa = 4.82STTGVNEE64 pKa = 4.35NIATFVLTADD74 pKa = 4.85RR75 pKa = 11.84IYY77 pKa = 11.02KK78 pKa = 10.31EE79 pKa = 4.32DD80 pKa = 3.37MKK82 pKa = 11.2FSIKK86 pKa = 10.44LNAAEE91 pKa = 4.28STADD95 pKa = 3.53AKK97 pKa = 11.28DD98 pKa = 3.62FTISLSEE105 pKa = 4.04ADD107 pKa = 4.69LDD109 pKa = 4.23YY110 pKa = 9.94GTDD113 pKa = 3.47GAFVATIPANTLSTTFTVAANFDD136 pKa = 3.95IYY138 pKa = 10.79PEE140 pKa = 4.15STEE143 pKa = 4.19NLVFDD148 pKa = 4.99IKK150 pKa = 11.07PMADD154 pKa = 3.4LNGTIAGSSKK164 pKa = 10.39KK165 pKa = 10.51VEE167 pKa = 3.97LTIGNSTSDD176 pKa = 3.97EE177 pKa = 4.15LVLILDD183 pKa = 3.83WNGDD187 pKa = 3.43STYY190 pKa = 11.49TNITDD195 pKa = 3.94PNDD198 pKa = 3.91PYY200 pKa = 11.41EE201 pKa = 4.44STLADD206 pKa = 3.45YY207 pKa = 11.32DD208 pKa = 4.13FDD210 pKa = 5.43LEE212 pKa = 4.65VYY214 pKa = 10.54NSGFSLVDD222 pKa = 3.5YY223 pKa = 10.07SWSDD227 pKa = 3.13SPEE230 pKa = 3.79QVNFATSNADD240 pKa = 2.88GLYY243 pKa = 9.95FVIAGFYY250 pKa = 8.07STYY253 pKa = 10.57GAVPMLPINFDD264 pKa = 3.45VTLTIAKK271 pKa = 9.43PGVFVYY277 pKa = 10.27DD278 pKa = 3.81YY279 pKa = 11.44DD280 pKa = 4.31MSGIWNSTDD289 pKa = 3.33GGDD292 pKa = 4.79DD293 pKa = 3.49EE294 pKa = 6.28GNVDD298 pKa = 4.06AYY300 pKa = 10.03QVPVYY305 pKa = 9.82FVKK308 pKa = 8.6TTTNGVATYY317 pKa = 9.73EE318 pKa = 4.29VYY320 pKa = 11.04NLADD324 pKa = 3.91DD325 pKa = 4.06TLLASGKK332 pKa = 10.38SSTDD336 pKa = 2.94LKK338 pKa = 11.37VYY340 pKa = 10.15FNSFKK345 pKa = 10.5RR346 pKa = 11.84VKK348 pKa = 10.42
Molecular weight: 38.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U8QVF3|A0A2U8QVF3_9FLAO DUF350 domain-containing protein OS=Flavobacterium sediminis OX=2201181 GN=DI487_08335 PE=3 SV=1
MM1 pKa = 7.46GGKK4 pKa = 9.29ALCFYY9 pKa = 10.64LFYY12 pKa = 11.18ARR14 pKa = 11.84WVWEE18 pKa = 3.98NKK20 pKa = 8.99RR21 pKa = 11.84VKK23 pKa = 9.58TMCLTRR29 pKa = 11.84GKK31 pKa = 10.33QGLSVGWKK39 pKa = 8.39TPLQGEE45 pKa = 4.37ARR47 pKa = 11.84TGRR50 pKa = 11.84AQRR53 pKa = 11.84TQRR56 pKa = 11.84SVPILGFGLDD66 pKa = 3.84FAA68 pKa = 6.63
MM1 pKa = 7.46GGKK4 pKa = 9.29ALCFYY9 pKa = 10.64LFYY12 pKa = 11.18ARR14 pKa = 11.84WVWEE18 pKa = 3.98NKK20 pKa = 8.99RR21 pKa = 11.84VKK23 pKa = 9.58TMCLTRR29 pKa = 11.84GKK31 pKa = 10.33QGLSVGWKK39 pKa = 8.39TPLQGEE45 pKa = 4.37ARR47 pKa = 11.84TGRR50 pKa = 11.84AQRR53 pKa = 11.84TQRR56 pKa = 11.84SVPILGFGLDD66 pKa = 3.84FAA68 pKa = 6.63
Molecular weight: 7.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
964564 |
28 |
3877 |
326.7 |
36.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.216 ± 0.042 | 0.812 ± 0.017 |
5.425 ± 0.039 | 6.837 ± 0.055 |
5.383 ± 0.045 | 6.19 ± 0.057 |
1.722 ± 0.022 | 7.834 ± 0.044 |
7.845 ± 0.073 | 9.149 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.136 ± 0.021 | 6.246 ± 0.05 |
3.312 ± 0.023 | 3.634 ± 0.026 |
3.148 ± 0.031 | 6.396 ± 0.047 |
6.023 ± 0.07 | 6.378 ± 0.036 |
1.006 ± 0.017 | 4.308 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
