
Pirellulimonas nuda
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Lacipirellulaceae; Pirellulimonas
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
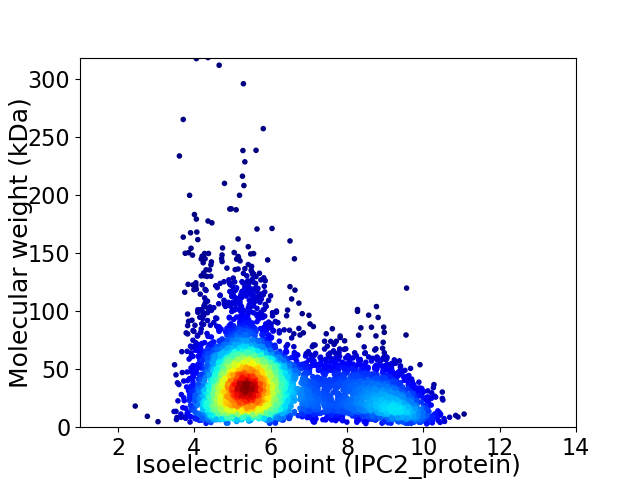
Virtual 2D-PAGE plot for 5103 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
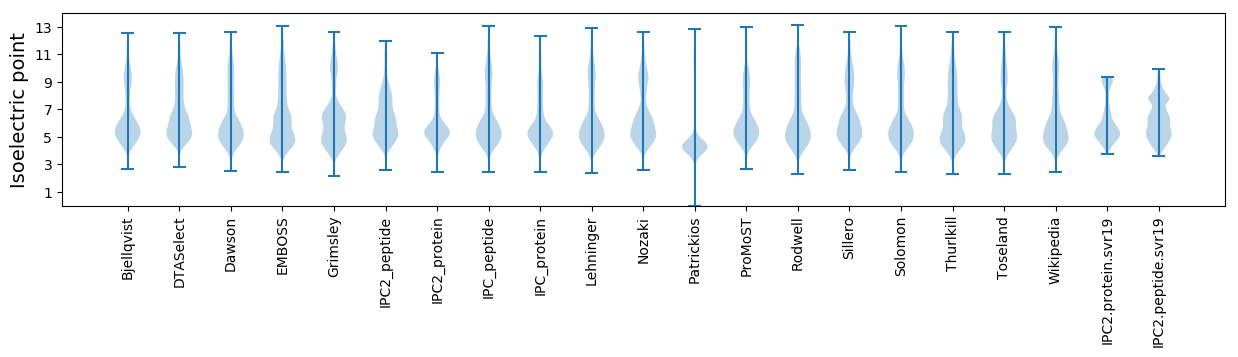
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A518DDH3|A0A518DDH3_9BACT Inositol 2-dehydrogenase OS=Pirellulimonas nuda OX=2528009 GN=iolG_7 PE=4 SV=1
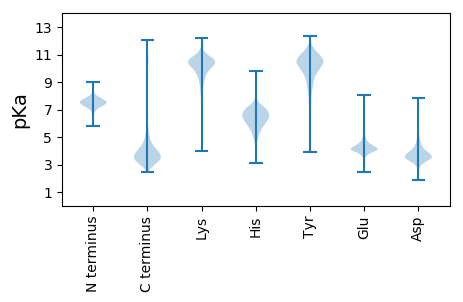
MM1 pKa = 7.73IFRR4 pKa = 11.84PYY6 pKa = 11.19ALTLALVALPGLSSAQSVLNIDD28 pKa = 4.18FGGGPNASGAGNFVGLAAYY47 pKa = 9.45GADD50 pKa = 3.68PAGAAALWNGINDD63 pKa = 4.32PLGDD67 pKa = 4.26AGTTNPLFDD76 pKa = 4.5SLNTPTSVSYY86 pKa = 10.89RR87 pKa = 11.84HH88 pKa = 5.03TTAGGGGTFIKK99 pKa = 10.22IPPAQEE105 pKa = 4.0VGTGDD110 pKa = 3.49PPQFLNLMNDD120 pKa = 3.99YY121 pKa = 10.68IWFNGGGTGRR131 pKa = 11.84TATISGLTPNATYY144 pKa = 11.06DD145 pKa = 3.78LYY147 pKa = 11.25AYY149 pKa = 9.76GQGDD153 pKa = 4.37TFTSLTSNQASKK165 pKa = 9.28FTVNGIEE172 pKa = 4.03KK173 pKa = 7.3TTTYY177 pKa = 9.82DD178 pKa = 3.01TDD180 pKa = 3.72AGGMLANDD188 pKa = 4.01GLLAEE193 pKa = 4.69NIEE196 pKa = 4.44YY197 pKa = 10.74VRR199 pKa = 11.84FTAVNSGPSGQIQISHH215 pKa = 5.97VNPTGGGFSAFNGLQIVGTFLAPPVPLNLEE245 pKa = 4.18VNTTNGTVTIQNNNQIPVQFDD266 pKa = 3.79VYY268 pKa = 10.99VIEE271 pKa = 4.45TSAGTLSKK279 pKa = 10.94SGWNSLSDD287 pKa = 3.56QNFDD291 pKa = 3.46ATAGAADD298 pKa = 4.25GDD300 pKa = 4.42YY301 pKa = 11.18NDD303 pKa = 5.58DD304 pKa = 3.99GAVDD308 pKa = 3.48AADD311 pKa = 3.51YY312 pKa = 8.25TVWRR316 pKa = 11.84DD317 pKa = 3.63SLNLPSAGNLEE328 pKa = 4.12NDD330 pKa = 4.71PIGGTVGAAQYY341 pKa = 9.61DD342 pKa = 3.48QWKK345 pKa = 10.52NGFGDD350 pKa = 3.69AGGTATGWDD359 pKa = 3.63EE360 pKa = 5.46AFGTSNSLLAEE371 pKa = 4.96FYY373 pKa = 10.64LGSGDD378 pKa = 4.32KK379 pKa = 9.89PASSIAAGQSLNLGPAFTSASADD402 pKa = 3.22GTLTFNYY409 pKa = 10.19YY410 pKa = 8.71DD411 pKa = 3.85TQASANMVGTVSFVNAAIGGNTVPEE436 pKa = 4.7PGCVWLLLAGAVSLGWRR453 pKa = 11.84LRR455 pKa = 11.84TPTNGRR461 pKa = 11.84CGG463 pKa = 3.35
MM1 pKa = 7.73IFRR4 pKa = 11.84PYY6 pKa = 11.19ALTLALVALPGLSSAQSVLNIDD28 pKa = 4.18FGGGPNASGAGNFVGLAAYY47 pKa = 9.45GADD50 pKa = 3.68PAGAAALWNGINDD63 pKa = 4.32PLGDD67 pKa = 4.26AGTTNPLFDD76 pKa = 4.5SLNTPTSVSYY86 pKa = 10.89RR87 pKa = 11.84HH88 pKa = 5.03TTAGGGGTFIKK99 pKa = 10.22IPPAQEE105 pKa = 4.0VGTGDD110 pKa = 3.49PPQFLNLMNDD120 pKa = 3.99YY121 pKa = 10.68IWFNGGGTGRR131 pKa = 11.84TATISGLTPNATYY144 pKa = 11.06DD145 pKa = 3.78LYY147 pKa = 11.25AYY149 pKa = 9.76GQGDD153 pKa = 4.37TFTSLTSNQASKK165 pKa = 9.28FTVNGIEE172 pKa = 4.03KK173 pKa = 7.3TTTYY177 pKa = 9.82DD178 pKa = 3.01TDD180 pKa = 3.72AGGMLANDD188 pKa = 4.01GLLAEE193 pKa = 4.69NIEE196 pKa = 4.44YY197 pKa = 10.74VRR199 pKa = 11.84FTAVNSGPSGQIQISHH215 pKa = 5.97VNPTGGGFSAFNGLQIVGTFLAPPVPLNLEE245 pKa = 4.18VNTTNGTVTIQNNNQIPVQFDD266 pKa = 3.79VYY268 pKa = 10.99VIEE271 pKa = 4.45TSAGTLSKK279 pKa = 10.94SGWNSLSDD287 pKa = 3.56QNFDD291 pKa = 3.46ATAGAADD298 pKa = 4.25GDD300 pKa = 4.42YY301 pKa = 11.18NDD303 pKa = 5.58DD304 pKa = 3.99GAVDD308 pKa = 3.48AADD311 pKa = 3.51YY312 pKa = 8.25TVWRR316 pKa = 11.84DD317 pKa = 3.63SLNLPSAGNLEE328 pKa = 4.12NDD330 pKa = 4.71PIGGTVGAAQYY341 pKa = 9.61DD342 pKa = 3.48QWKK345 pKa = 10.52NGFGDD350 pKa = 3.69AGGTATGWDD359 pKa = 3.63EE360 pKa = 5.46AFGTSNSLLAEE371 pKa = 4.96FYY373 pKa = 10.64LGSGDD378 pKa = 4.32KK379 pKa = 9.89PASSIAAGQSLNLGPAFTSASADD402 pKa = 3.22GTLTFNYY409 pKa = 10.19YY410 pKa = 8.71DD411 pKa = 3.85TQASANMVGTVSFVNAAIGGNTVPEE436 pKa = 4.7PGCVWLLLAGAVSLGWRR453 pKa = 11.84LRR455 pKa = 11.84TPTNGRR461 pKa = 11.84CGG463 pKa = 3.35
Molecular weight: 47.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A518DCP5|A0A518DCP5_9BACT PEP-CTERM domain-containing protein OS=Pirellulimonas nuda OX=2528009 GN=Pla175_26390 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84FAAASPEE9 pKa = 4.13PEE11 pKa = 4.05SLICRR16 pKa = 11.84SVQVEE21 pKa = 4.4DD22 pKa = 4.57GVATLLVEE30 pKa = 4.61SGRR33 pKa = 11.84QTAHH37 pKa = 6.7CPSCGEE43 pKa = 3.75PSRR46 pKa = 11.84RR47 pKa = 11.84VHH49 pKa = 6.04SRR51 pKa = 11.84YY52 pKa = 9.6VRR54 pKa = 11.84NLKK57 pKa = 10.17DD58 pKa = 3.9LSWHH62 pKa = 5.84GLQVRR67 pKa = 11.84LVWRR71 pKa = 11.84SRR73 pKa = 11.84KK74 pKa = 9.43LFCRR78 pKa = 11.84NAGCQQRR85 pKa = 11.84VFTEE89 pKa = 4.35RR90 pKa = 11.84LPKK93 pKa = 9.89VAQPHH98 pKa = 5.72ARR100 pKa = 11.84QTTRR104 pKa = 11.84SEE106 pKa = 3.7EE107 pKa = 4.01ALRR110 pKa = 11.84AIAMACGGLPGARR123 pKa = 11.84PANRR127 pKa = 11.84LGMATSGDD135 pKa = 3.58SLLRR139 pKa = 11.84LLRR142 pKa = 11.84RR143 pKa = 11.84AGHH146 pKa = 6.52KK147 pKa = 10.32KK148 pKa = 8.78PACPQVLGVDD158 pKa = 4.68DD159 pKa = 3.44IAFRR163 pKa = 11.84KK164 pKa = 9.12RR165 pKa = 11.84ATYY168 pKa = 8.23GTILCDD174 pKa = 3.43LVRR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84PIHH182 pKa = 6.26LLPEE186 pKa = 4.18RR187 pKa = 11.84SKK189 pKa = 11.3EE190 pKa = 4.12SLSAWLSKK198 pKa = 10.76HH199 pKa = 6.04SGVKK203 pKa = 9.98IISRR207 pKa = 11.84DD208 pKa = 2.98RR209 pKa = 11.84GEE211 pKa = 4.26YY212 pKa = 8.7YY213 pKa = 10.33RR214 pKa = 11.84QGASEE219 pKa = 4.29GAPHH223 pKa = 7.46ALQVADD229 pKa = 4.75RR230 pKa = 11.84WHH232 pKa = 6.59LLCNLRR238 pKa = 11.84EE239 pKa = 4.1ALVRR243 pKa = 11.84WLGRR247 pKa = 11.84CSRR250 pKa = 11.84EE251 pKa = 3.73WPMDD255 pKa = 3.31
MM1 pKa = 7.62RR2 pKa = 11.84FAAASPEE9 pKa = 4.13PEE11 pKa = 4.05SLICRR16 pKa = 11.84SVQVEE21 pKa = 4.4DD22 pKa = 4.57GVATLLVEE30 pKa = 4.61SGRR33 pKa = 11.84QTAHH37 pKa = 6.7CPSCGEE43 pKa = 3.75PSRR46 pKa = 11.84RR47 pKa = 11.84VHH49 pKa = 6.04SRR51 pKa = 11.84YY52 pKa = 9.6VRR54 pKa = 11.84NLKK57 pKa = 10.17DD58 pKa = 3.9LSWHH62 pKa = 5.84GLQVRR67 pKa = 11.84LVWRR71 pKa = 11.84SRR73 pKa = 11.84KK74 pKa = 9.43LFCRR78 pKa = 11.84NAGCQQRR85 pKa = 11.84VFTEE89 pKa = 4.35RR90 pKa = 11.84LPKK93 pKa = 9.89VAQPHH98 pKa = 5.72ARR100 pKa = 11.84QTTRR104 pKa = 11.84SEE106 pKa = 3.7EE107 pKa = 4.01ALRR110 pKa = 11.84AIAMACGGLPGARR123 pKa = 11.84PANRR127 pKa = 11.84LGMATSGDD135 pKa = 3.58SLLRR139 pKa = 11.84LLRR142 pKa = 11.84RR143 pKa = 11.84AGHH146 pKa = 6.52KK147 pKa = 10.32KK148 pKa = 8.78PACPQVLGVDD158 pKa = 4.68DD159 pKa = 3.44IAFRR163 pKa = 11.84KK164 pKa = 9.12RR165 pKa = 11.84ATYY168 pKa = 8.23GTILCDD174 pKa = 3.43LVRR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84PIHH182 pKa = 6.26LLPEE186 pKa = 4.18RR187 pKa = 11.84SKK189 pKa = 11.3EE190 pKa = 4.12SLSAWLSKK198 pKa = 10.76HH199 pKa = 6.04SGVKK203 pKa = 9.98IISRR207 pKa = 11.84DD208 pKa = 2.98RR209 pKa = 11.84GEE211 pKa = 4.26YY212 pKa = 8.7YY213 pKa = 10.33RR214 pKa = 11.84QGASEE219 pKa = 4.29GAPHH223 pKa = 7.46ALQVADD229 pKa = 4.75RR230 pKa = 11.84WHH232 pKa = 6.59LLCNLRR238 pKa = 11.84EE239 pKa = 4.1ALVRR243 pKa = 11.84WLGRR247 pKa = 11.84CSRR250 pKa = 11.84EE251 pKa = 3.73WPMDD255 pKa = 3.31
Molecular weight: 28.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1869859 |
29 |
3095 |
366.4 |
39.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.064 ± 0.052 | 1.117 ± 0.014 |
6.064 ± 0.029 | 5.814 ± 0.029 |
3.376 ± 0.018 | 8.85 ± 0.038 |
2.016 ± 0.015 | 3.905 ± 0.024 |
2.805 ± 0.03 | 9.991 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.969 ± 0.014 | 2.714 ± 0.028 |
5.839 ± 0.028 | 3.718 ± 0.022 |
7.237 ± 0.038 | 5.798 ± 0.029 |
5.217 ± 0.029 | 7.527 ± 0.027 |
1.574 ± 0.014 | 2.404 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
