
Lawsonia intracellularis (strain PHE/MN1-00)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Lawsonia; Lawsonia intracellularis
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
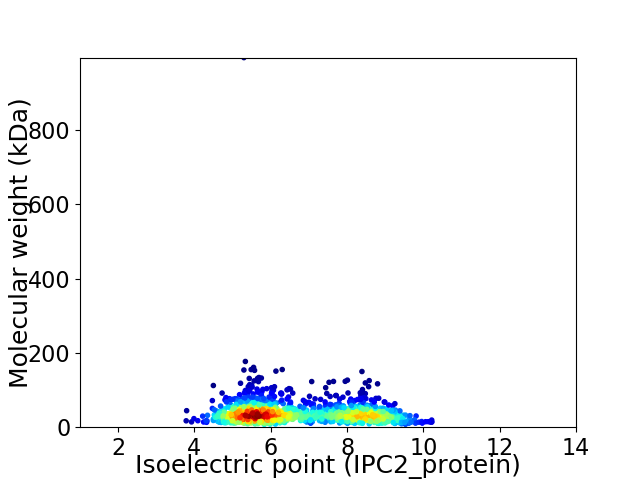
Virtual 2D-PAGE plot for 1342 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
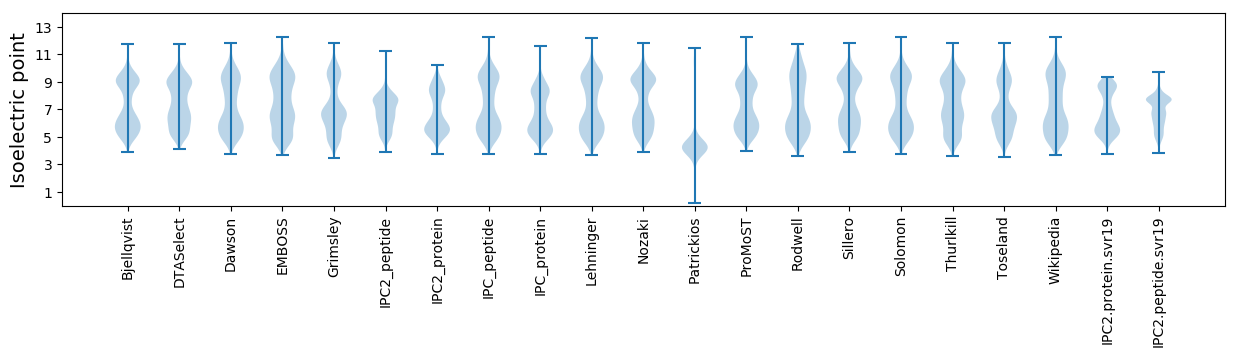
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1MSG0|Q1MSG0_LAWIP NA OS=Lawsonia intracellularis (strain PHE/MN1-00) OX=363253 GN=LI0009 PE=4 SV=1
MM1 pKa = 7.39VSTDD5 pKa = 3.48VIDD8 pKa = 4.72ALGHH12 pKa = 6.34LLNIPGLSFNEE23 pKa = 4.43DD24 pKa = 4.08LLCLLSVNNDD34 pKa = 3.15NVDD37 pKa = 3.43VTIEE41 pKa = 4.23TNEE44 pKa = 4.21SGDD47 pKa = 3.77LLYY50 pKa = 11.29VCAHH54 pKa = 7.38LGTMPDD60 pKa = 4.13DD61 pKa = 4.99DD62 pKa = 4.16NAAAAIALLFAQANAVLTAMNKK84 pKa = 9.46GVLVLDD90 pKa = 4.22YY91 pKa = 11.02NGQGLMLVKK100 pKa = 10.21LYY102 pKa = 10.71EE103 pKa = 4.12MKK105 pKa = 10.23HH106 pKa = 5.36WEE108 pKa = 4.86LEE110 pKa = 4.16DD111 pKa = 3.76CLEE114 pKa = 5.66SILQLFNDD122 pKa = 3.27AGEE125 pKa = 4.36WKK127 pKa = 10.31NRR129 pKa = 11.84LYY131 pKa = 11.15LPDD134 pKa = 4.72FGLGGMDD141 pKa = 4.45FDD143 pKa = 6.59LDD145 pKa = 4.02TSSSGSDD152 pKa = 2.84QFMRR156 pKa = 11.84VV157 pKa = 2.94
MM1 pKa = 7.39VSTDD5 pKa = 3.48VIDD8 pKa = 4.72ALGHH12 pKa = 6.34LLNIPGLSFNEE23 pKa = 4.43DD24 pKa = 4.08LLCLLSVNNDD34 pKa = 3.15NVDD37 pKa = 3.43VTIEE41 pKa = 4.23TNEE44 pKa = 4.21SGDD47 pKa = 3.77LLYY50 pKa = 11.29VCAHH54 pKa = 7.38LGTMPDD60 pKa = 4.13DD61 pKa = 4.99DD62 pKa = 4.16NAAAAIALLFAQANAVLTAMNKK84 pKa = 9.46GVLVLDD90 pKa = 4.22YY91 pKa = 11.02NGQGLMLVKK100 pKa = 10.21LYY102 pKa = 10.71EE103 pKa = 4.12MKK105 pKa = 10.23HH106 pKa = 5.36WEE108 pKa = 4.86LEE110 pKa = 4.16DD111 pKa = 3.76CLEE114 pKa = 5.66SILQLFNDD122 pKa = 3.27AGEE125 pKa = 4.36WKK127 pKa = 10.31NRR129 pKa = 11.84LYY131 pKa = 11.15LPDD134 pKa = 4.72FGLGGMDD141 pKa = 4.45FDD143 pKa = 6.59LDD145 pKa = 4.02TSSSGSDD152 pKa = 2.84QFMRR156 pKa = 11.84VV157 pKa = 2.94
Molecular weight: 17.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q1MPR3|RL3_LAWIP 50S ribosomal protein L3 OS=Lawsonia intracellularis (strain PHE/MN1-00) OX=363253 GN=rplC PE=3 SV=1
MM1 pKa = 8.13AIRR4 pKa = 11.84NLKK7 pKa = 7.94PTSPGCRR14 pKa = 11.84FQTISDD20 pKa = 3.93FKK22 pKa = 10.93EE23 pKa = 4.17ITCSSPEE30 pKa = 3.63KK31 pKa = 10.49SLTYY35 pKa = 10.23GLCKK39 pKa = 10.07KK40 pKa = 10.48AGRR43 pKa = 11.84NNHH46 pKa = 4.71GHH48 pKa = 4.93ITSRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84GGGAKK59 pKa = 9.66RR60 pKa = 11.84LYY62 pKa = 10.55RR63 pKa = 11.84LIDD66 pKa = 3.81FKK68 pKa = 10.88RR69 pKa = 11.84DD70 pKa = 3.45KK71 pKa = 11.11VGVPAKK77 pKa = 10.02VAQIEE82 pKa = 4.15YY83 pKa = 10.53DD84 pKa = 3.53PNRR87 pKa = 11.84TARR90 pKa = 11.84IALLHH95 pKa = 5.76YY96 pKa = 10.51VDD98 pKa = 3.9GEE100 pKa = 3.97KK101 pKa = 10.21RR102 pKa = 11.84YY103 pKa = 9.94ILAPVGIKK111 pKa = 10.07QGDD114 pKa = 3.81IIEE117 pKa = 4.61TGVDD121 pKa = 3.27ADD123 pKa = 3.83IKK125 pKa = 10.4PGNSLPMLHH134 pKa = 7.02IPIGTLIHH142 pKa = 6.67NIEE145 pKa = 4.46LNPGRR150 pKa = 11.84GGQLCRR156 pKa = 11.84SAGAYY161 pKa = 8.29AQLVARR167 pKa = 11.84EE168 pKa = 4.21GKK170 pKa = 9.91YY171 pKa = 10.76ALLRR175 pKa = 11.84MPSGEE180 pKa = 3.95VRR182 pKa = 11.84KK183 pKa = 10.22VLASGAATVGQVGNIHH199 pKa = 6.77HH200 pKa = 6.65EE201 pKa = 4.39NISLGKK207 pKa = 10.13AGRR210 pKa = 11.84NRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PKK219 pKa = 10.49VRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.73HH230 pKa = 7.25PLGGGEE236 pKa = 4.15GRR238 pKa = 11.84SSGGRR243 pKa = 11.84HH244 pKa = 5.52PVSPWGMPTKK254 pKa = 10.4GYY256 pKa = 7.8KK257 pKa = 9.22TRR259 pKa = 11.84DD260 pKa = 3.2KK261 pKa = 10.64KK262 pKa = 11.12KK263 pKa = 8.6PSSRR267 pKa = 11.84LIVKK271 pKa = 10.17RR272 pKa = 11.84RR273 pKa = 11.84GQKK276 pKa = 9.78
MM1 pKa = 8.13AIRR4 pKa = 11.84NLKK7 pKa = 7.94PTSPGCRR14 pKa = 11.84FQTISDD20 pKa = 3.93FKK22 pKa = 10.93EE23 pKa = 4.17ITCSSPEE30 pKa = 3.63KK31 pKa = 10.49SLTYY35 pKa = 10.23GLCKK39 pKa = 10.07KK40 pKa = 10.48AGRR43 pKa = 11.84NNHH46 pKa = 4.71GHH48 pKa = 4.93ITSRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84GGGAKK59 pKa = 9.66RR60 pKa = 11.84LYY62 pKa = 10.55RR63 pKa = 11.84LIDD66 pKa = 3.81FKK68 pKa = 10.88RR69 pKa = 11.84DD70 pKa = 3.45KK71 pKa = 11.11VGVPAKK77 pKa = 10.02VAQIEE82 pKa = 4.15YY83 pKa = 10.53DD84 pKa = 3.53PNRR87 pKa = 11.84TARR90 pKa = 11.84IALLHH95 pKa = 5.76YY96 pKa = 10.51VDD98 pKa = 3.9GEE100 pKa = 3.97KK101 pKa = 10.21RR102 pKa = 11.84YY103 pKa = 9.94ILAPVGIKK111 pKa = 10.07QGDD114 pKa = 3.81IIEE117 pKa = 4.61TGVDD121 pKa = 3.27ADD123 pKa = 3.83IKK125 pKa = 10.4PGNSLPMLHH134 pKa = 7.02IPIGTLIHH142 pKa = 6.67NIEE145 pKa = 4.46LNPGRR150 pKa = 11.84GGQLCRR156 pKa = 11.84SAGAYY161 pKa = 8.29AQLVARR167 pKa = 11.84EE168 pKa = 4.21GKK170 pKa = 9.91YY171 pKa = 10.76ALLRR175 pKa = 11.84MPSGEE180 pKa = 3.95VRR182 pKa = 11.84KK183 pKa = 10.22VLASGAATVGQVGNIHH199 pKa = 6.77HH200 pKa = 6.65EE201 pKa = 4.39NISLGKK207 pKa = 10.13AGRR210 pKa = 11.84NRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PKK219 pKa = 10.49VRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.73HH230 pKa = 7.25PLGGGEE236 pKa = 4.15GRR238 pKa = 11.84SSGGRR243 pKa = 11.84HH244 pKa = 5.52PVSPWGMPTKK254 pKa = 10.4GYY256 pKa = 7.8KK257 pKa = 9.22TRR259 pKa = 11.84DD260 pKa = 3.2KK261 pKa = 10.64KK262 pKa = 11.12KK263 pKa = 8.6PSSRR267 pKa = 11.84LIVKK271 pKa = 10.17RR272 pKa = 11.84RR273 pKa = 11.84GQKK276 pKa = 9.78
Molecular weight: 30.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
474690 |
67 |
8746 |
353.7 |
39.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.336 ± 0.073 | 1.349 ± 0.024 |
4.436 ± 0.055 | 6.247 ± 0.165 |
4.328 ± 0.068 | 6.463 ± 0.064 |
2.306 ± 0.033 | 8.599 ± 0.126 |
6.332 ± 0.123 | 10.602 ± 0.106 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.212 ± 0.036 | 4.622 ± 0.06 |
4.349 ± 0.046 | 4.193 ± 0.082 |
4.087 ± 0.049 | 7.243 ± 0.099 |
5.959 ± 0.05 | 5.837 ± 0.056 |
1.147 ± 0.028 | 3.353 ± 0.049 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
