
Roseivivax isoporae LMG 25204
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseivivax; Roseivivax isoporae
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
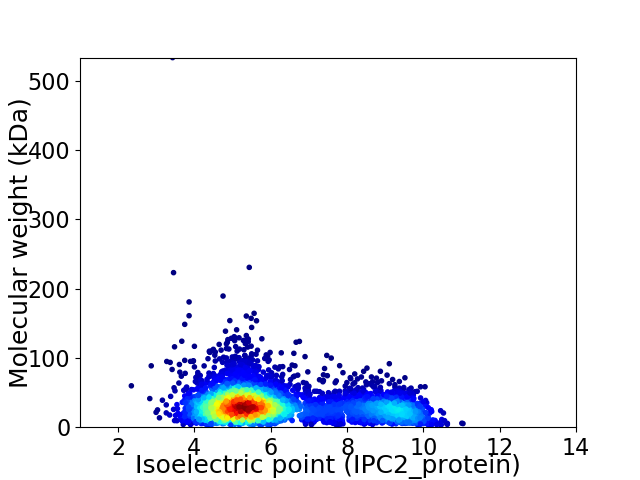
Virtual 2D-PAGE plot for 4626 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
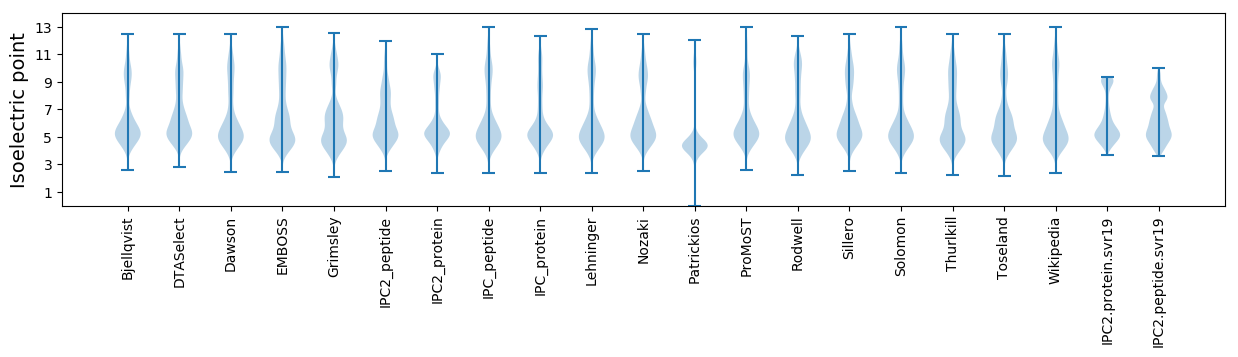
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|X7F3A6|X7F3A6_9RHOB Cation:proton antiporter OS=Roseivivax isoporae LMG 25204 OX=1449351 GN=RISW2_13910 PE=3 SV=1
MM1 pKa = 7.24NRR3 pKa = 11.84TAAIALAAALAAVPAVAQQSGEE25 pKa = 4.27SLSGSLVLYY34 pKa = 8.97TSQPDD39 pKa = 2.99QDD41 pKa = 3.7AQQTVDD47 pKa = 4.3AFNEE51 pKa = 4.34VYY53 pKa = 10.52PDD55 pKa = 3.96IDD57 pKa = 3.98VEE59 pKa = 4.3WVRR62 pKa = 11.84DD63 pKa = 3.58GTTQMMARR71 pKa = 11.84LRR73 pKa = 11.84AEE75 pKa = 4.42FEE77 pKa = 4.19AGQPQPDD84 pKa = 3.71VLLIADD90 pKa = 4.56TVTMEE95 pKa = 4.35SLEE98 pKa = 4.12QEE100 pKa = 4.58GRR102 pKa = 11.84LMAYY106 pKa = 9.06TDD108 pKa = 4.44ADD110 pKa = 3.43VSAYY114 pKa = 10.41DD115 pKa = 4.0EE116 pKa = 4.62GLMDD120 pKa = 3.89PEE122 pKa = 5.21GYY124 pKa = 10.53YY125 pKa = 10.8FSTKK129 pKa = 10.53LITTGIVYY137 pKa = 10.59NEE139 pKa = 4.15AAEE142 pKa = 4.27MVPQSYY148 pKa = 11.59ADD150 pKa = 3.91LLEE153 pKa = 4.38PAAEE157 pKa = 4.21NNIVMPSPLEE167 pKa = 4.14SGAATIHH174 pKa = 5.55MVSLTGLPEE183 pKa = 4.64LGWDD187 pKa = 4.07YY188 pKa = 11.54YY189 pKa = 11.25QGLADD194 pKa = 4.21QGAIAQGGNGGTYY207 pKa = 9.3QAVAGGQALYY217 pKa = 10.97GFVVDD222 pKa = 7.14FLALRR227 pKa = 11.84NIQDD231 pKa = 3.68GAPVGFVFPEE241 pKa = 4.09EE242 pKa = 4.17GVSAVTEE249 pKa = 3.97PVAILEE255 pKa = 4.23TARR258 pKa = 11.84NVEE261 pKa = 3.92AAQAFVDD268 pKa = 4.39FLLSEE273 pKa = 4.89RR274 pKa = 11.84GQEE277 pKa = 3.61LAAAQGYY284 pKa = 9.47LPAHH288 pKa = 7.25PDD290 pKa = 3.08VAAPDD295 pKa = 3.95YY296 pKa = 10.93FPDD299 pKa = 3.7RR300 pKa = 11.84STIKK304 pKa = 10.94VIDD307 pKa = 4.02FDD309 pKa = 4.24PAEE312 pKa = 4.31ALANDD317 pKa = 3.75AAYY320 pKa = 9.76KK321 pKa = 10.42EE322 pKa = 4.48RR323 pKa = 11.84FAEE326 pKa = 4.32TFGGG330 pKa = 3.75
MM1 pKa = 7.24NRR3 pKa = 11.84TAAIALAAALAAVPAVAQQSGEE25 pKa = 4.27SLSGSLVLYY34 pKa = 8.97TSQPDD39 pKa = 2.99QDD41 pKa = 3.7AQQTVDD47 pKa = 4.3AFNEE51 pKa = 4.34VYY53 pKa = 10.52PDD55 pKa = 3.96IDD57 pKa = 3.98VEE59 pKa = 4.3WVRR62 pKa = 11.84DD63 pKa = 3.58GTTQMMARR71 pKa = 11.84LRR73 pKa = 11.84AEE75 pKa = 4.42FEE77 pKa = 4.19AGQPQPDD84 pKa = 3.71VLLIADD90 pKa = 4.56TVTMEE95 pKa = 4.35SLEE98 pKa = 4.12QEE100 pKa = 4.58GRR102 pKa = 11.84LMAYY106 pKa = 9.06TDD108 pKa = 4.44ADD110 pKa = 3.43VSAYY114 pKa = 10.41DD115 pKa = 4.0EE116 pKa = 4.62GLMDD120 pKa = 3.89PEE122 pKa = 5.21GYY124 pKa = 10.53YY125 pKa = 10.8FSTKK129 pKa = 10.53LITTGIVYY137 pKa = 10.59NEE139 pKa = 4.15AAEE142 pKa = 4.27MVPQSYY148 pKa = 11.59ADD150 pKa = 3.91LLEE153 pKa = 4.38PAAEE157 pKa = 4.21NNIVMPSPLEE167 pKa = 4.14SGAATIHH174 pKa = 5.55MVSLTGLPEE183 pKa = 4.64LGWDD187 pKa = 4.07YY188 pKa = 11.54YY189 pKa = 11.25QGLADD194 pKa = 4.21QGAIAQGGNGGTYY207 pKa = 9.3QAVAGGQALYY217 pKa = 10.97GFVVDD222 pKa = 7.14FLALRR227 pKa = 11.84NIQDD231 pKa = 3.68GAPVGFVFPEE241 pKa = 4.09EE242 pKa = 4.17GVSAVTEE249 pKa = 3.97PVAILEE255 pKa = 4.23TARR258 pKa = 11.84NVEE261 pKa = 3.92AAQAFVDD268 pKa = 4.39FLLSEE273 pKa = 4.89RR274 pKa = 11.84GQEE277 pKa = 3.61LAAAQGYY284 pKa = 9.47LPAHH288 pKa = 7.25PDD290 pKa = 3.08VAAPDD295 pKa = 3.95YY296 pKa = 10.93FPDD299 pKa = 3.7RR300 pKa = 11.84STIKK304 pKa = 10.94VIDD307 pKa = 4.02FDD309 pKa = 4.24PAEE312 pKa = 4.31ALANDD317 pKa = 3.75AAYY320 pKa = 9.76KK321 pKa = 10.42EE322 pKa = 4.48RR323 pKa = 11.84FAEE326 pKa = 4.32TFGGG330 pKa = 3.75
Molecular weight: 35.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X7F8R9|X7F8R9_9RHOB Uncharacterized protein OS=Roseivivax isoporae LMG 25204 OX=1449351 GN=RISW2_06215 PE=4 SV=1
MM1 pKa = 7.5SIKK4 pKa = 10.86GIVTKK9 pKa = 9.85VAKK12 pKa = 10.32GYY14 pKa = 10.35ARR16 pKa = 11.84SKK18 pKa = 9.8SGHH21 pKa = 5.07GAARR25 pKa = 11.84RR26 pKa = 11.84PVNRR30 pKa = 11.84PVRR33 pKa = 11.84RR34 pKa = 11.84TGGKK38 pKa = 8.26GQLVRR43 pKa = 11.84GLLNFARR50 pKa = 11.84KK51 pKa = 9.19KK52 pKa = 9.12VV53 pKa = 3.34
MM1 pKa = 7.5SIKK4 pKa = 10.86GIVTKK9 pKa = 9.85VAKK12 pKa = 10.32GYY14 pKa = 10.35ARR16 pKa = 11.84SKK18 pKa = 9.8SGHH21 pKa = 5.07GAARR25 pKa = 11.84RR26 pKa = 11.84PVNRR30 pKa = 11.84PVRR33 pKa = 11.84RR34 pKa = 11.84TGGKK38 pKa = 8.26GQLVRR43 pKa = 11.84GLLNFARR50 pKa = 11.84KK51 pKa = 9.19KK52 pKa = 9.12VV53 pKa = 3.34
Molecular weight: 5.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1446104 |
28 |
5196 |
312.6 |
33.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.849 ± 0.063 | 0.829 ± 0.012 |
6.347 ± 0.036 | 5.828 ± 0.034 |
3.512 ± 0.023 | 9.253 ± 0.045 |
1.996 ± 0.019 | 4.466 ± 0.028 |
2.037 ± 0.026 | 10.187 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.548 ± 0.017 | 2.082 ± 0.021 |
5.369 ± 0.024 | 2.693 ± 0.019 |
7.862 ± 0.043 | 4.619 ± 0.024 |
5.437 ± 0.026 | 7.653 ± 0.03 |
1.403 ± 0.015 | 2.029 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
