
Azoarcus sp. CC-YHH838
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Rhodocyclales; Zoogloeaceae; Azoarcus
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
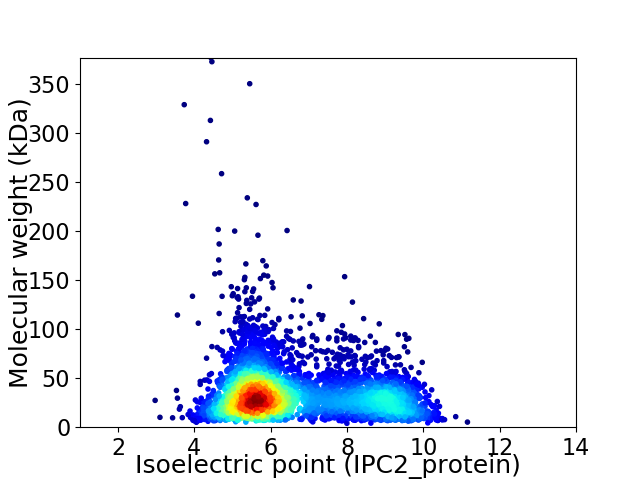
Virtual 2D-PAGE plot for 4235 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S4AWT3|A0A4S4AWT3_9RHOO AbrB family transcriptional regulator OS=Azoarcus sp. CC-YHH838 OX=2565930 GN=E6C76_10610 PE=4 SV=1
TT1 pKa = 6.9ATATDD6 pKa = 3.79NASNTATDD14 pKa = 3.8TDD16 pKa = 3.88TAGYY20 pKa = 10.53DD21 pKa = 3.5LLPTVGVSFEE31 pKa = 4.03ISDD34 pKa = 3.81AGVGTLTGTSEE45 pKa = 4.18HH46 pKa = 6.03ATSVSVTITGSDD58 pKa = 3.57GSNLGPFPATLNPDD72 pKa = 3.62GSWSVDD78 pKa = 3.32LSGEE82 pKa = 4.21SFDD85 pKa = 5.45EE86 pKa = 4.29EE87 pKa = 4.25VTYY90 pKa = 9.34TATATATDD98 pKa = 3.88NASNTATGSDD108 pKa = 3.21TAGYY112 pKa = 10.53DD113 pKa = 3.51LLPTVGVSFEE123 pKa = 4.03ISDD126 pKa = 3.81AGVGTLTGTSEE137 pKa = 4.18HH138 pKa = 6.03ATSVSVTITGSDD150 pKa = 3.57GSNLGPFPATLNPDD164 pKa = 3.62GSWSVDD170 pKa = 3.32LSGEE174 pKa = 4.21SFDD177 pKa = 5.45EE178 pKa = 4.29EE179 pKa = 4.25VTYY182 pKa = 9.34TATATATDD190 pKa = 3.88NASNTATDD198 pKa = 3.8TDD200 pKa = 3.88TAGYY204 pKa = 10.53DD205 pKa = 3.5LLPTVGVSFEE215 pKa = 4.03ISDD218 pKa = 3.81AGVGTLTGTSEE229 pKa = 4.18HH230 pKa = 6.03ATSVSVTITGSDD242 pKa = 3.57GSNLGPFPATLNPDD256 pKa = 3.62GTWSVDD262 pKa = 3.44LSGEE266 pKa = 4.21SFDD269 pKa = 5.45EE270 pKa = 4.29EE271 pKa = 4.25VTYY274 pKa = 9.34TATATATDD282 pKa = 3.88NASNTATGSDD292 pKa = 3.31TASYY296 pKa = 10.68DD297 pKa = 3.58LNAPPVAIDD306 pKa = 4.49DD307 pKa = 4.24PTGQPYY313 pKa = 10.25SVSLGSLGSGWSNPDD328 pKa = 3.47SNHH331 pKa = 6.52SLTDD335 pKa = 3.78LSAWKK340 pKa = 10.49ADD342 pKa = 3.42GTAGEE347 pKa = 5.3LYY349 pKa = 10.43QSGDD353 pKa = 3.22MRR355 pKa = 11.84GVKK358 pKa = 10.35GSPRR362 pKa = 11.84GGALDD367 pKa = 3.81QEE369 pKa = 4.65PDD371 pKa = 3.28QLQSNNSGQSEE382 pKa = 4.38AIEE385 pKa = 4.26LKK387 pKa = 10.81FSGNLDD393 pKa = 3.17QATFSVSHH401 pKa = 6.94LYY403 pKa = 11.03SNEE406 pKa = 3.68NGYY409 pKa = 10.9GEE411 pKa = 4.02IGRR414 pKa = 11.84WVAYY418 pKa = 9.99FDD420 pKa = 3.94GVEE423 pKa = 4.13VASGEE428 pKa = 4.2FRR430 pKa = 11.84FNDD433 pKa = 3.81LTNSTHH439 pKa = 6.64SGTFAIDD446 pKa = 3.04TGGRR450 pKa = 11.84LFNSVRR456 pKa = 11.84FEE458 pKa = 3.95AVHH461 pKa = 6.41NGDD464 pKa = 3.61SSGDD468 pKa = 3.42SSGYY472 pKa = 10.29FLTGFAGTGPEE483 pKa = 4.32VANGTYY489 pKa = 9.98LVSVGDD495 pKa = 3.76TLNVSEE501 pKa = 5.69LEE503 pKa = 4.73GILANDD509 pKa = 4.04SDD511 pKa = 4.73PEE513 pKa = 4.1NDD515 pKa = 3.74PLSIVAVNGSALTVGEE531 pKa = 4.79WITLPSGAKK540 pKa = 10.32LMLNADD546 pKa = 3.78GSFSYY551 pKa = 9.19DD552 pKa = 3.0TNNQFGHH559 pKa = 6.84LAAGEE564 pKa = 4.05WATDD568 pKa = 3.26SFQYY572 pKa = 10.29TISDD576 pKa = 3.5GMGGEE581 pKa = 4.41DD582 pKa = 3.44TATVTVTVVGTGVPVDD598 pKa = 3.55TSVTDD603 pKa = 3.83GNEE606 pKa = 4.28VISTAEE612 pKa = 3.99DD613 pKa = 3.26AVLTGNVLANTDD625 pKa = 3.99TNVGTPAVSSFSVAGDD641 pKa = 3.34PAIYY645 pKa = 10.19DD646 pKa = 3.88AGDD649 pKa = 3.21TATISGVGTLTIGADD664 pKa = 3.22GEE666 pKa = 4.73YY667 pKa = 10.15RR668 pKa = 11.84FEE670 pKa = 4.2PAQDD674 pKa = 3.48YY675 pKa = 10.66SGPVPTVTYY684 pKa = 9.31TVTNGVQSDD693 pKa = 3.85TSTLEE698 pKa = 3.72ITVTPVADD706 pKa = 3.68QPEE709 pKa = 4.43VSVDD713 pKa = 3.46VQVAGSVVQTLTTANVLGTPTGFTVTAYY741 pKa = 10.02KK742 pKa = 10.67DD743 pKa = 3.68GEE745 pKa = 4.19PATLSVRR752 pKa = 11.84SSGSPTGFGVAGSASDD768 pKa = 3.78GASEE772 pKa = 4.32EE773 pKa = 4.15IGNGEE778 pKa = 4.24VLRR781 pKa = 11.84VQLQTAAEE789 pKa = 4.29SVSFRR794 pKa = 11.84LAWLASGEE802 pKa = 4.15YY803 pKa = 10.19AKK805 pKa = 10.98YY806 pKa = 10.63VLIYY810 pKa = 10.42TDD812 pKa = 3.8GTSEE816 pKa = 4.56SVILNGNSAVGGYY829 pKa = 9.65DD830 pKa = 3.04QVGSVITRR838 pKa = 11.84NAPAGKK844 pKa = 9.72LIAAIEE850 pKa = 4.24FTTPGVGEE858 pKa = 3.71AHH860 pKa = 6.02YY861 pKa = 10.85AGNNDD866 pKa = 3.33YY867 pKa = 11.1LVYY870 pKa = 10.15QVSYY874 pKa = 10.45VAASSYY880 pKa = 10.33EE881 pKa = 4.06LNIVATPTDD890 pKa = 3.62NYY892 pKa = 8.53HH893 pKa = 6.08TEE895 pKa = 4.35SITEE899 pKa = 4.05LVVVVPVGITLSAGTHH915 pKa = 6.44DD916 pKa = 5.84GSGKK920 pKa = 7.4WTLPLDD926 pKa = 3.74GAGAYY931 pKa = 7.23TVQVDD936 pKa = 3.96PVTKK940 pKa = 9.17EE941 pKa = 3.79VSVSGLSMNVPSDD954 pKa = 3.5FSGEE958 pKa = 3.94LTLTVTAKK966 pKa = 10.74AADD969 pKa = 3.89GTDD972 pKa = 3.32TAEE975 pKa = 4.8GSTSLNIEE983 pKa = 4.18RR984 pKa = 11.84GTAGNDD990 pKa = 3.43TLVGTDD996 pKa = 3.84GADD999 pKa = 3.15ILIGGAGDD1007 pKa = 4.01DD1008 pKa = 4.08VLTGGLGADD1017 pKa = 3.36VFVWRR1022 pKa = 11.84LGDD1025 pKa = 3.34QGTNSQPATDD1035 pKa = 4.02RR1036 pKa = 11.84VTDD1039 pKa = 4.01FNLGQGDD1046 pKa = 4.1VLDD1049 pKa = 5.66LRR1051 pKa = 11.84DD1052 pKa = 4.44LLDD1055 pKa = 3.71GHH1057 pKa = 6.68SGSLDD1062 pKa = 3.07HH1063 pKa = 6.37YY1064 pKa = 10.68LKK1066 pKa = 10.85VEE1068 pKa = 4.04EE1069 pKa = 4.52SSGNTVLTISTTGNVNASHH1088 pKa = 6.84DD1089 pKa = 3.88QVIVLDD1095 pKa = 4.4GVTGLIDD1102 pKa = 4.38GGLTSQSAIIQDD1114 pKa = 5.1LINKK1118 pKa = 8.08QAIQIDD1124 pKa = 3.74
TT1 pKa = 6.9ATATDD6 pKa = 3.79NASNTATDD14 pKa = 3.8TDD16 pKa = 3.88TAGYY20 pKa = 10.53DD21 pKa = 3.5LLPTVGVSFEE31 pKa = 4.03ISDD34 pKa = 3.81AGVGTLTGTSEE45 pKa = 4.18HH46 pKa = 6.03ATSVSVTITGSDD58 pKa = 3.57GSNLGPFPATLNPDD72 pKa = 3.62GSWSVDD78 pKa = 3.32LSGEE82 pKa = 4.21SFDD85 pKa = 5.45EE86 pKa = 4.29EE87 pKa = 4.25VTYY90 pKa = 9.34TATATATDD98 pKa = 3.88NASNTATGSDD108 pKa = 3.21TAGYY112 pKa = 10.53DD113 pKa = 3.51LLPTVGVSFEE123 pKa = 4.03ISDD126 pKa = 3.81AGVGTLTGTSEE137 pKa = 4.18HH138 pKa = 6.03ATSVSVTITGSDD150 pKa = 3.57GSNLGPFPATLNPDD164 pKa = 3.62GSWSVDD170 pKa = 3.32LSGEE174 pKa = 4.21SFDD177 pKa = 5.45EE178 pKa = 4.29EE179 pKa = 4.25VTYY182 pKa = 9.34TATATATDD190 pKa = 3.88NASNTATDD198 pKa = 3.8TDD200 pKa = 3.88TAGYY204 pKa = 10.53DD205 pKa = 3.5LLPTVGVSFEE215 pKa = 4.03ISDD218 pKa = 3.81AGVGTLTGTSEE229 pKa = 4.18HH230 pKa = 6.03ATSVSVTITGSDD242 pKa = 3.57GSNLGPFPATLNPDD256 pKa = 3.62GTWSVDD262 pKa = 3.44LSGEE266 pKa = 4.21SFDD269 pKa = 5.45EE270 pKa = 4.29EE271 pKa = 4.25VTYY274 pKa = 9.34TATATATDD282 pKa = 3.88NASNTATGSDD292 pKa = 3.31TASYY296 pKa = 10.68DD297 pKa = 3.58LNAPPVAIDD306 pKa = 4.49DD307 pKa = 4.24PTGQPYY313 pKa = 10.25SVSLGSLGSGWSNPDD328 pKa = 3.47SNHH331 pKa = 6.52SLTDD335 pKa = 3.78LSAWKK340 pKa = 10.49ADD342 pKa = 3.42GTAGEE347 pKa = 5.3LYY349 pKa = 10.43QSGDD353 pKa = 3.22MRR355 pKa = 11.84GVKK358 pKa = 10.35GSPRR362 pKa = 11.84GGALDD367 pKa = 3.81QEE369 pKa = 4.65PDD371 pKa = 3.28QLQSNNSGQSEE382 pKa = 4.38AIEE385 pKa = 4.26LKK387 pKa = 10.81FSGNLDD393 pKa = 3.17QATFSVSHH401 pKa = 6.94LYY403 pKa = 11.03SNEE406 pKa = 3.68NGYY409 pKa = 10.9GEE411 pKa = 4.02IGRR414 pKa = 11.84WVAYY418 pKa = 9.99FDD420 pKa = 3.94GVEE423 pKa = 4.13VASGEE428 pKa = 4.2FRR430 pKa = 11.84FNDD433 pKa = 3.81LTNSTHH439 pKa = 6.64SGTFAIDD446 pKa = 3.04TGGRR450 pKa = 11.84LFNSVRR456 pKa = 11.84FEE458 pKa = 3.95AVHH461 pKa = 6.41NGDD464 pKa = 3.61SSGDD468 pKa = 3.42SSGYY472 pKa = 10.29FLTGFAGTGPEE483 pKa = 4.32VANGTYY489 pKa = 9.98LVSVGDD495 pKa = 3.76TLNVSEE501 pKa = 5.69LEE503 pKa = 4.73GILANDD509 pKa = 4.04SDD511 pKa = 4.73PEE513 pKa = 4.1NDD515 pKa = 3.74PLSIVAVNGSALTVGEE531 pKa = 4.79WITLPSGAKK540 pKa = 10.32LMLNADD546 pKa = 3.78GSFSYY551 pKa = 9.19DD552 pKa = 3.0TNNQFGHH559 pKa = 6.84LAAGEE564 pKa = 4.05WATDD568 pKa = 3.26SFQYY572 pKa = 10.29TISDD576 pKa = 3.5GMGGEE581 pKa = 4.41DD582 pKa = 3.44TATVTVTVVGTGVPVDD598 pKa = 3.55TSVTDD603 pKa = 3.83GNEE606 pKa = 4.28VISTAEE612 pKa = 3.99DD613 pKa = 3.26AVLTGNVLANTDD625 pKa = 3.99TNVGTPAVSSFSVAGDD641 pKa = 3.34PAIYY645 pKa = 10.19DD646 pKa = 3.88AGDD649 pKa = 3.21TATISGVGTLTIGADD664 pKa = 3.22GEE666 pKa = 4.73YY667 pKa = 10.15RR668 pKa = 11.84FEE670 pKa = 4.2PAQDD674 pKa = 3.48YY675 pKa = 10.66SGPVPTVTYY684 pKa = 9.31TVTNGVQSDD693 pKa = 3.85TSTLEE698 pKa = 3.72ITVTPVADD706 pKa = 3.68QPEE709 pKa = 4.43VSVDD713 pKa = 3.46VQVAGSVVQTLTTANVLGTPTGFTVTAYY741 pKa = 10.02KK742 pKa = 10.67DD743 pKa = 3.68GEE745 pKa = 4.19PATLSVRR752 pKa = 11.84SSGSPTGFGVAGSASDD768 pKa = 3.78GASEE772 pKa = 4.32EE773 pKa = 4.15IGNGEE778 pKa = 4.24VLRR781 pKa = 11.84VQLQTAAEE789 pKa = 4.29SVSFRR794 pKa = 11.84LAWLASGEE802 pKa = 4.15YY803 pKa = 10.19AKK805 pKa = 10.98YY806 pKa = 10.63VLIYY810 pKa = 10.42TDD812 pKa = 3.8GTSEE816 pKa = 4.56SVILNGNSAVGGYY829 pKa = 9.65DD830 pKa = 3.04QVGSVITRR838 pKa = 11.84NAPAGKK844 pKa = 9.72LIAAIEE850 pKa = 4.24FTTPGVGEE858 pKa = 3.71AHH860 pKa = 6.02YY861 pKa = 10.85AGNNDD866 pKa = 3.33YY867 pKa = 11.1LVYY870 pKa = 10.15QVSYY874 pKa = 10.45VAASSYY880 pKa = 10.33EE881 pKa = 4.06LNIVATPTDD890 pKa = 3.62NYY892 pKa = 8.53HH893 pKa = 6.08TEE895 pKa = 4.35SITEE899 pKa = 4.05LVVVVPVGITLSAGTHH915 pKa = 6.44DD916 pKa = 5.84GSGKK920 pKa = 7.4WTLPLDD926 pKa = 3.74GAGAYY931 pKa = 7.23TVQVDD936 pKa = 3.96PVTKK940 pKa = 9.17EE941 pKa = 3.79VSVSGLSMNVPSDD954 pKa = 3.5FSGEE958 pKa = 3.94LTLTVTAKK966 pKa = 10.74AADD969 pKa = 3.89GTDD972 pKa = 3.32TAEE975 pKa = 4.8GSTSLNIEE983 pKa = 4.18RR984 pKa = 11.84GTAGNDD990 pKa = 3.43TLVGTDD996 pKa = 3.84GADD999 pKa = 3.15ILIGGAGDD1007 pKa = 4.01DD1008 pKa = 4.08VLTGGLGADD1017 pKa = 3.36VFVWRR1022 pKa = 11.84LGDD1025 pKa = 3.34QGTNSQPATDD1035 pKa = 4.02RR1036 pKa = 11.84VTDD1039 pKa = 4.01FNLGQGDD1046 pKa = 4.1VLDD1049 pKa = 5.66LRR1051 pKa = 11.84DD1052 pKa = 4.44LLDD1055 pKa = 3.71GHH1057 pKa = 6.68SGSLDD1062 pKa = 3.07HH1063 pKa = 6.37YY1064 pKa = 10.68LKK1066 pKa = 10.85VEE1068 pKa = 4.04EE1069 pKa = 4.52SSGNTVLTISTTGNVNASHH1088 pKa = 6.84DD1089 pKa = 3.88QVIVLDD1095 pKa = 4.4GVTGLIDD1102 pKa = 4.38GGLTSQSAIIQDD1114 pKa = 5.1LINKK1118 pKa = 8.08QAIQIDD1124 pKa = 3.74
Molecular weight: 114.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S4AYU7|A0A4S4AYU7_9RHOO Integrating conjugative element protein OS=Azoarcus sp. CC-YHH838 OX=2565930 GN=E6C76_12590 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.75GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.75GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1419603 |
35 |
3709 |
335.2 |
36.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.182 ± 0.057 | 0.929 ± 0.013 |
5.419 ± 0.026 | 5.918 ± 0.035 |
3.468 ± 0.023 | 8.587 ± 0.051 |
2.312 ± 0.021 | 4.323 ± 0.029 |
2.579 ± 0.026 | 11.242 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.103 ± 0.019 | 2.509 ± 0.027 |
5.153 ± 0.034 | 3.51 ± 0.023 |
7.92 ± 0.046 | 4.958 ± 0.035 |
4.855 ± 0.042 | 7.344 ± 0.031 |
1.411 ± 0.018 | 2.28 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
