
Streptomyces sp. DSM 15324
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
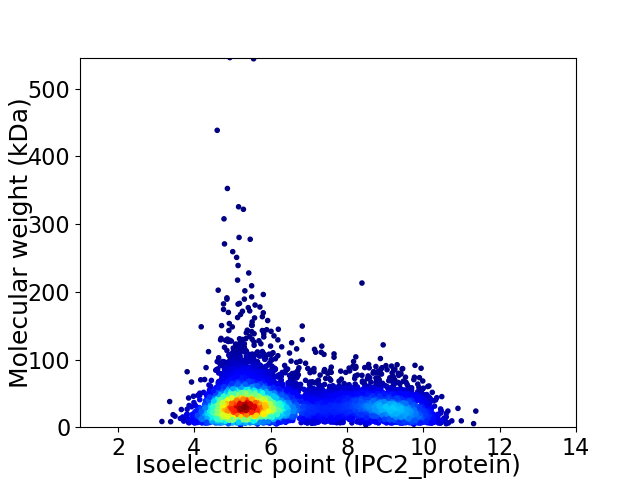
Virtual 2D-PAGE plot for 7596 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A101UJI4|A0A101UJI4_9ACTN LuxR family transcriptional regulator OS=Streptomyces sp. DSM 15324 OX=1739111 GN=AQJ58_11835 PE=4 SV=1
MM1 pKa = 7.81SIPPPPGSPQPQDD14 pKa = 3.75PYY16 pKa = 10.94RR17 pKa = 11.84PPPPHH22 pKa = 6.68GPYY25 pKa = 9.86PQGPYY30 pKa = 9.24PQAPQAPYY38 pKa = 10.23GAVPGPYY45 pKa = 9.14QVWGQGYY52 pKa = 9.65SPLARR57 pKa = 11.84PAPVNGVAIAALVLGLLCFLPAVGLILGIIALVQIKK93 pKa = 10.33KK94 pKa = 10.08RR95 pKa = 11.84GEE97 pKa = 3.94RR98 pKa = 11.84GKK100 pKa = 11.49GMAIAGAVVSSLGLALWALSLSTNVASEE128 pKa = 3.77FWEE131 pKa = 4.88GVKK134 pKa = 10.44EE135 pKa = 4.05GARR138 pKa = 11.84GGGYY142 pKa = 10.08SLAAGDD148 pKa = 4.67CFDD151 pKa = 4.38VPGSFDD157 pKa = 4.07RR158 pKa = 11.84DD159 pKa = 3.64VYY161 pKa = 11.47DD162 pKa = 3.55VDD164 pKa = 4.94EE165 pKa = 4.98VPCSDD170 pKa = 3.39PHH172 pKa = 7.5EE173 pKa = 4.91GEE175 pKa = 4.25AFGTIPLSGDD185 pKa = 3.61DD186 pKa = 4.19YY187 pKa = 11.49PGDD190 pKa = 4.16GYY192 pKa = 10.48VTDD195 pKa = 3.92QAEE198 pKa = 4.64DD199 pKa = 3.24KK200 pKa = 11.04CGPLQDD206 pKa = 4.48AYY208 pKa = 11.1AMDD211 pKa = 3.97RR212 pKa = 11.84WALGDD217 pKa = 3.88EE218 pKa = 4.08VDD220 pKa = 4.58VYY222 pKa = 11.65YY223 pKa = 9.37LTPTADD229 pKa = 2.76SWEE232 pKa = 4.06WGDD235 pKa = 5.2RR236 pKa = 11.84EE237 pKa = 4.28ITCVFAHH244 pKa = 6.29TDD246 pKa = 3.31EE247 pKa = 4.89TGTLTGSLRR256 pKa = 11.84ADD258 pKa = 3.76EE259 pKa = 4.48TTLDD263 pKa = 3.8ADD265 pKa = 3.75QMAFLKK271 pKa = 10.91AMAAIDD277 pKa = 3.61DD278 pKa = 4.11VLYY281 pKa = 10.74EE282 pKa = 4.36EE283 pKa = 5.35PDD285 pKa = 3.82DD286 pKa = 4.08YY287 pKa = 12.17AEE289 pKa = 5.63ADD291 pKa = 3.44LDD293 pKa = 4.92ANKK296 pKa = 10.59DD297 pKa = 2.98WAADD301 pKa = 3.58VRR303 pKa = 11.84DD304 pKa = 3.98VLAEE308 pKa = 3.76QAGALDD314 pKa = 3.5AHH316 pKa = 6.3TWSGDD321 pKa = 3.04AGKK324 pKa = 10.5SVDD327 pKa = 3.35ALVRR331 pKa = 11.84EE332 pKa = 4.36MRR334 pKa = 11.84DD335 pKa = 4.04ARR337 pKa = 11.84ADD339 pKa = 3.24WAKK342 pKa = 10.84AATAGDD348 pKa = 3.85ADD350 pKa = 4.11TFYY353 pKa = 11.54LHH355 pKa = 6.69YY356 pKa = 9.89EE357 pKa = 3.7SGYY360 pKa = 11.11AYY362 pKa = 10.94VDD364 pKa = 3.21GDD366 pKa = 3.88TTVAARR372 pKa = 11.84KK373 pKa = 9.67ALDD376 pKa = 3.7LATTPPSYY384 pKa = 11.2DD385 pKa = 3.45EE386 pKa = 5.57DD387 pKa = 3.96EE388 pKa = 4.95GDD390 pKa = 4.28GGDD393 pKa = 3.73SGSGSGSGSGSGGSGSEE410 pKa = 4.34DD411 pKa = 3.24VV412 pKa = 4.05
MM1 pKa = 7.81SIPPPPGSPQPQDD14 pKa = 3.75PYY16 pKa = 10.94RR17 pKa = 11.84PPPPHH22 pKa = 6.68GPYY25 pKa = 9.86PQGPYY30 pKa = 9.24PQAPQAPYY38 pKa = 10.23GAVPGPYY45 pKa = 9.14QVWGQGYY52 pKa = 9.65SPLARR57 pKa = 11.84PAPVNGVAIAALVLGLLCFLPAVGLILGIIALVQIKK93 pKa = 10.33KK94 pKa = 10.08RR95 pKa = 11.84GEE97 pKa = 3.94RR98 pKa = 11.84GKK100 pKa = 11.49GMAIAGAVVSSLGLALWALSLSTNVASEE128 pKa = 3.77FWEE131 pKa = 4.88GVKK134 pKa = 10.44EE135 pKa = 4.05GARR138 pKa = 11.84GGGYY142 pKa = 10.08SLAAGDD148 pKa = 4.67CFDD151 pKa = 4.38VPGSFDD157 pKa = 4.07RR158 pKa = 11.84DD159 pKa = 3.64VYY161 pKa = 11.47DD162 pKa = 3.55VDD164 pKa = 4.94EE165 pKa = 4.98VPCSDD170 pKa = 3.39PHH172 pKa = 7.5EE173 pKa = 4.91GEE175 pKa = 4.25AFGTIPLSGDD185 pKa = 3.61DD186 pKa = 4.19YY187 pKa = 11.49PGDD190 pKa = 4.16GYY192 pKa = 10.48VTDD195 pKa = 3.92QAEE198 pKa = 4.64DD199 pKa = 3.24KK200 pKa = 11.04CGPLQDD206 pKa = 4.48AYY208 pKa = 11.1AMDD211 pKa = 3.97RR212 pKa = 11.84WALGDD217 pKa = 3.88EE218 pKa = 4.08VDD220 pKa = 4.58VYY222 pKa = 11.65YY223 pKa = 9.37LTPTADD229 pKa = 2.76SWEE232 pKa = 4.06WGDD235 pKa = 5.2RR236 pKa = 11.84EE237 pKa = 4.28ITCVFAHH244 pKa = 6.29TDD246 pKa = 3.31EE247 pKa = 4.89TGTLTGSLRR256 pKa = 11.84ADD258 pKa = 3.76EE259 pKa = 4.48TTLDD263 pKa = 3.8ADD265 pKa = 3.75QMAFLKK271 pKa = 10.91AMAAIDD277 pKa = 3.61DD278 pKa = 4.11VLYY281 pKa = 10.74EE282 pKa = 4.36EE283 pKa = 5.35PDD285 pKa = 3.82DD286 pKa = 4.08YY287 pKa = 12.17AEE289 pKa = 5.63ADD291 pKa = 3.44LDD293 pKa = 4.92ANKK296 pKa = 10.59DD297 pKa = 2.98WAADD301 pKa = 3.58VRR303 pKa = 11.84DD304 pKa = 3.98VLAEE308 pKa = 3.76QAGALDD314 pKa = 3.5AHH316 pKa = 6.3TWSGDD321 pKa = 3.04AGKK324 pKa = 10.5SVDD327 pKa = 3.35ALVRR331 pKa = 11.84EE332 pKa = 4.36MRR334 pKa = 11.84DD335 pKa = 4.04ARR337 pKa = 11.84ADD339 pKa = 3.24WAKK342 pKa = 10.84AATAGDD348 pKa = 3.85ADD350 pKa = 4.11TFYY353 pKa = 11.54LHH355 pKa = 6.69YY356 pKa = 9.89EE357 pKa = 3.7SGYY360 pKa = 11.11AYY362 pKa = 10.94VDD364 pKa = 3.21GDD366 pKa = 3.88TTVAARR372 pKa = 11.84KK373 pKa = 9.67ALDD376 pKa = 3.7LATTPPSYY384 pKa = 11.2DD385 pKa = 3.45EE386 pKa = 5.57DD387 pKa = 3.96EE388 pKa = 4.95GDD390 pKa = 4.28GGDD393 pKa = 3.73SGSGSGSGSGSGGSGSEE410 pKa = 4.34DD411 pKa = 3.24VV412 pKa = 4.05
Molecular weight: 43.19 kDa
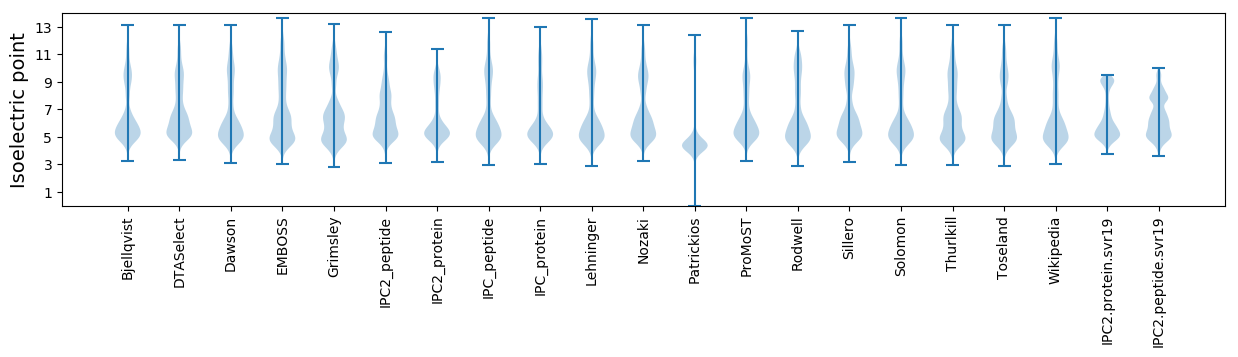
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A101UDP8|A0A101UDP8_9ACTN DNA polymerase III subunit gamma/tau OS=Streptomyces sp. DSM 15324 OX=1739111 GN=AQJ58_29525 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.75GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.75GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2550604 |
29 |
5257 |
335.8 |
35.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.651 ± 0.045 | 0.793 ± 0.007 |
6.014 ± 0.022 | 5.62 ± 0.032 |
2.714 ± 0.016 | 9.511 ± 0.029 |
2.34 ± 0.016 | 3.001 ± 0.018 |
2.056 ± 0.023 | 10.45 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.646 ± 0.012 | 1.715 ± 0.016 |
6.127 ± 0.029 | 2.703 ± 0.016 |
8.144 ± 0.035 | 4.999 ± 0.018 |
6.28 ± 0.026 | 8.601 ± 0.024 |
1.52 ± 0.012 | 2.114 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
