
Marinilactibacillus sp. 15R
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Carnobacteriaceae; Marinilactibacillus; unclassified Marinilactibacillus
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
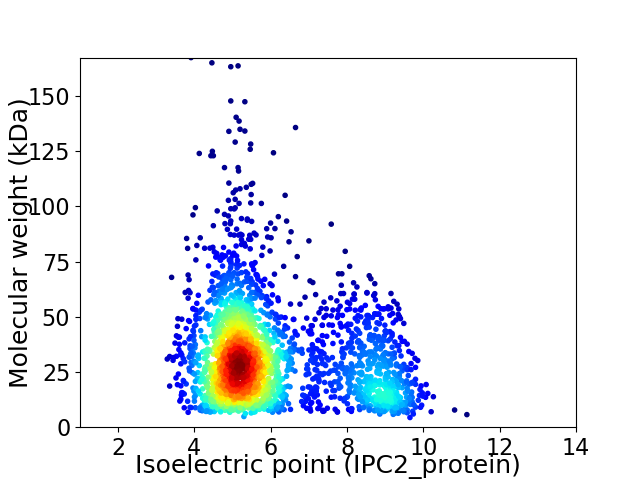
Virtual 2D-PAGE plot for 2486 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L4BZL0|A0A1L4BZL0_9LACT Dihydroorotase OS=Marinilactibacillus sp. 15R OX=1911586 GN=pyrC PE=3 SV=1
MM1 pKa = 7.45SNKK4 pKa = 9.73KK5 pKa = 10.09KK6 pKa = 10.73YY7 pKa = 10.59SLLASSAAVAFVLAACGGSDD27 pKa = 5.14NADD30 pKa = 3.45SDD32 pKa = 4.4NGSSDD37 pKa = 3.3NGGSEE42 pKa = 4.26SGSEE46 pKa = 4.23GPLTFEE52 pKa = 3.97TRR54 pKa = 11.84VEE56 pKa = 4.18NEE58 pKa = 3.77GEE60 pKa = 4.21AIEE63 pKa = 4.8GGTMNIGYY71 pKa = 10.2VSDD74 pKa = 3.98SPFTGIFNWEE84 pKa = 4.33LYY86 pKa = 10.46DD87 pKa = 4.22GNPDD91 pKa = 3.71AQVMAYY97 pKa = 9.54TMGEE101 pKa = 3.97LVGSDD106 pKa = 3.5EE107 pKa = 4.94NYY109 pKa = 10.71QIDD112 pKa = 4.06DD113 pKa = 3.67SGAATLEE120 pKa = 4.02LDD122 pKa = 3.6QEE124 pKa = 4.74ANTVTITVKK133 pKa = 10.87DD134 pKa = 4.03GIKK137 pKa = 9.22WHH139 pKa = 7.12DD140 pKa = 4.17GEE142 pKa = 5.31PLTADD147 pKa = 3.44DD148 pKa = 4.55VLFAHH153 pKa = 7.28EE154 pKa = 4.83IIGDD158 pKa = 3.71PEE160 pKa = 4.33YY161 pKa = 11.02TGVRR165 pKa = 11.84YY166 pKa = 7.47TADD169 pKa = 3.31LQNIVGMEE177 pKa = 3.97EE178 pKa = 4.35YY179 pKa = 10.84KK180 pKa = 11.01NGEE183 pKa = 3.99ADD185 pKa = 4.03SISGMTLSDD194 pKa = 4.54DD195 pKa = 3.88EE196 pKa = 4.47MTLTIQYY203 pKa = 10.33KK204 pKa = 9.75SVGVQMLQAGGGVWTYY220 pKa = 11.67AEE222 pKa = 4.14PRR224 pKa = 11.84HH225 pKa = 5.53YY226 pKa = 11.05LGDD229 pKa = 3.6IPVADD234 pKa = 4.76LEE236 pKa = 4.52SAPEE240 pKa = 4.01VRR242 pKa = 11.84EE243 pKa = 3.94KK244 pKa = 10.85PLGYY248 pKa = 10.89GPFKK252 pKa = 10.65VDD254 pKa = 3.48NIVPGEE260 pKa = 4.19SVSYY264 pKa = 11.15SRR266 pKa = 11.84FDD268 pKa = 3.78DD269 pKa = 3.71YY270 pKa = 11.6WRR272 pKa = 11.84GTPILDD278 pKa = 4.45GIEE281 pKa = 4.17LQTVPTATAVAAMEE295 pKa = 4.16NAQFDD300 pKa = 4.61LFIDD304 pKa = 4.09MPTDD308 pKa = 3.3QYY310 pKa = 11.75DD311 pKa = 3.59TFADD315 pKa = 3.9LPGYY319 pKa = 10.17SIVGRR324 pKa = 11.84DD325 pKa = 3.69EE326 pKa = 4.3NSYY329 pKa = 9.83TYY331 pKa = 10.35IGFKK335 pKa = 10.31LGEE338 pKa = 4.01WQEE341 pKa = 3.98AQEE344 pKa = 4.49DD345 pKa = 4.17DD346 pKa = 4.12EE347 pKa = 5.86GNITEE352 pKa = 4.3PAKK355 pKa = 10.72NVYY358 pKa = 10.42NPDD361 pKa = 2.81AKK363 pKa = 10.52MADD366 pKa = 3.42KK367 pKa = 10.86NLRR370 pKa = 11.84QAMAYY375 pKa = 10.12AIDD378 pKa = 3.78NDD380 pKa = 4.01AVGEE384 pKa = 4.04RR385 pKa = 11.84FYY387 pKa = 11.23QGLRR391 pKa = 11.84RR392 pKa = 11.84RR393 pKa = 11.84ANSPIIPNFSDD404 pKa = 4.28YY405 pKa = 11.47YY406 pKa = 10.19NEE408 pKa = 4.65DD409 pKa = 3.61VVGYY413 pKa = 9.22PYY415 pKa = 11.04DD416 pKa = 3.77EE417 pKa = 5.73DD418 pKa = 3.65EE419 pKa = 4.82ANRR422 pKa = 11.84ILDD425 pKa = 3.51EE426 pKa = 5.55AGYY429 pKa = 9.65EE430 pKa = 3.99WAEE433 pKa = 4.17GEE435 pKa = 4.41DD436 pKa = 4.21FRR438 pKa = 11.84TTPEE442 pKa = 4.08GEE444 pKa = 3.76EE445 pKa = 3.88LVINFASMSGGDD457 pKa = 3.54VAEE460 pKa = 5.66PIAEE464 pKa = 4.17YY465 pKa = 10.69YY466 pKa = 7.98IQSWARR472 pKa = 11.84IGLNVQLLEE481 pKa = 4.44GRR483 pKa = 11.84LHH485 pKa = 6.47EE486 pKa = 4.93FNSFYY491 pKa = 11.31DD492 pKa = 3.47RR493 pKa = 11.84VEE495 pKa = 4.92ADD497 pKa = 4.09DD498 pKa = 4.21PAIDD502 pKa = 4.13VYY504 pKa = 10.97QGAWGTGTDD513 pKa = 3.78PTPEE517 pKa = 3.87GLYY520 pKa = 11.14GEE522 pKa = 4.45TAPFNYY528 pKa = 8.66TRR530 pKa = 11.84WVTEE534 pKa = 3.7EE535 pKa = 3.62NNEE538 pKa = 3.9YY539 pKa = 9.2MEE541 pKa = 4.44QMLSSEE547 pKa = 4.4GFDD550 pKa = 3.6TEE552 pKa = 3.53WRR554 pKa = 11.84AEE556 pKa = 4.18VFNEE560 pKa = 3.42WQEE563 pKa = 4.2YY564 pKa = 9.52FMEE567 pKa = 4.71EE568 pKa = 4.2LPIIPTLFRR577 pKa = 11.84TEE579 pKa = 4.04VFPVNDD585 pKa = 3.27RR586 pKa = 11.84VGAFDD591 pKa = 3.2ITVGVDD597 pKa = 3.15PEE599 pKa = 4.31TAGAEE604 pKa = 4.29TGYY607 pKa = 8.56HH608 pKa = 4.1TWYY611 pKa = 8.88LTSEE615 pKa = 4.18EE616 pKa = 4.72RR617 pKa = 11.84VTEE620 pKa = 4.11
MM1 pKa = 7.45SNKK4 pKa = 9.73KK5 pKa = 10.09KK6 pKa = 10.73YY7 pKa = 10.59SLLASSAAVAFVLAACGGSDD27 pKa = 5.14NADD30 pKa = 3.45SDD32 pKa = 4.4NGSSDD37 pKa = 3.3NGGSEE42 pKa = 4.26SGSEE46 pKa = 4.23GPLTFEE52 pKa = 3.97TRR54 pKa = 11.84VEE56 pKa = 4.18NEE58 pKa = 3.77GEE60 pKa = 4.21AIEE63 pKa = 4.8GGTMNIGYY71 pKa = 10.2VSDD74 pKa = 3.98SPFTGIFNWEE84 pKa = 4.33LYY86 pKa = 10.46DD87 pKa = 4.22GNPDD91 pKa = 3.71AQVMAYY97 pKa = 9.54TMGEE101 pKa = 3.97LVGSDD106 pKa = 3.5EE107 pKa = 4.94NYY109 pKa = 10.71QIDD112 pKa = 4.06DD113 pKa = 3.67SGAATLEE120 pKa = 4.02LDD122 pKa = 3.6QEE124 pKa = 4.74ANTVTITVKK133 pKa = 10.87DD134 pKa = 4.03GIKK137 pKa = 9.22WHH139 pKa = 7.12DD140 pKa = 4.17GEE142 pKa = 5.31PLTADD147 pKa = 3.44DD148 pKa = 4.55VLFAHH153 pKa = 7.28EE154 pKa = 4.83IIGDD158 pKa = 3.71PEE160 pKa = 4.33YY161 pKa = 11.02TGVRR165 pKa = 11.84YY166 pKa = 7.47TADD169 pKa = 3.31LQNIVGMEE177 pKa = 3.97EE178 pKa = 4.35YY179 pKa = 10.84KK180 pKa = 11.01NGEE183 pKa = 3.99ADD185 pKa = 4.03SISGMTLSDD194 pKa = 4.54DD195 pKa = 3.88EE196 pKa = 4.47MTLTIQYY203 pKa = 10.33KK204 pKa = 9.75SVGVQMLQAGGGVWTYY220 pKa = 11.67AEE222 pKa = 4.14PRR224 pKa = 11.84HH225 pKa = 5.53YY226 pKa = 11.05LGDD229 pKa = 3.6IPVADD234 pKa = 4.76LEE236 pKa = 4.52SAPEE240 pKa = 4.01VRR242 pKa = 11.84EE243 pKa = 3.94KK244 pKa = 10.85PLGYY248 pKa = 10.89GPFKK252 pKa = 10.65VDD254 pKa = 3.48NIVPGEE260 pKa = 4.19SVSYY264 pKa = 11.15SRR266 pKa = 11.84FDD268 pKa = 3.78DD269 pKa = 3.71YY270 pKa = 11.6WRR272 pKa = 11.84GTPILDD278 pKa = 4.45GIEE281 pKa = 4.17LQTVPTATAVAAMEE295 pKa = 4.16NAQFDD300 pKa = 4.61LFIDD304 pKa = 4.09MPTDD308 pKa = 3.3QYY310 pKa = 11.75DD311 pKa = 3.59TFADD315 pKa = 3.9LPGYY319 pKa = 10.17SIVGRR324 pKa = 11.84DD325 pKa = 3.69EE326 pKa = 4.3NSYY329 pKa = 9.83TYY331 pKa = 10.35IGFKK335 pKa = 10.31LGEE338 pKa = 4.01WQEE341 pKa = 3.98AQEE344 pKa = 4.49DD345 pKa = 4.17DD346 pKa = 4.12EE347 pKa = 5.86GNITEE352 pKa = 4.3PAKK355 pKa = 10.72NVYY358 pKa = 10.42NPDD361 pKa = 2.81AKK363 pKa = 10.52MADD366 pKa = 3.42KK367 pKa = 10.86NLRR370 pKa = 11.84QAMAYY375 pKa = 10.12AIDD378 pKa = 3.78NDD380 pKa = 4.01AVGEE384 pKa = 4.04RR385 pKa = 11.84FYY387 pKa = 11.23QGLRR391 pKa = 11.84RR392 pKa = 11.84RR393 pKa = 11.84ANSPIIPNFSDD404 pKa = 4.28YY405 pKa = 11.47YY406 pKa = 10.19NEE408 pKa = 4.65DD409 pKa = 3.61VVGYY413 pKa = 9.22PYY415 pKa = 11.04DD416 pKa = 3.77EE417 pKa = 5.73DD418 pKa = 3.65EE419 pKa = 4.82ANRR422 pKa = 11.84ILDD425 pKa = 3.51EE426 pKa = 5.55AGYY429 pKa = 9.65EE430 pKa = 3.99WAEE433 pKa = 4.17GEE435 pKa = 4.41DD436 pKa = 4.21FRR438 pKa = 11.84TTPEE442 pKa = 4.08GEE444 pKa = 3.76EE445 pKa = 3.88LVINFASMSGGDD457 pKa = 3.54VAEE460 pKa = 5.66PIAEE464 pKa = 4.17YY465 pKa = 10.69YY466 pKa = 7.98IQSWARR472 pKa = 11.84IGLNVQLLEE481 pKa = 4.44GRR483 pKa = 11.84LHH485 pKa = 6.47EE486 pKa = 4.93FNSFYY491 pKa = 11.31DD492 pKa = 3.47RR493 pKa = 11.84VEE495 pKa = 4.92ADD497 pKa = 4.09DD498 pKa = 4.21PAIDD502 pKa = 4.13VYY504 pKa = 10.97QGAWGTGTDD513 pKa = 3.78PTPEE517 pKa = 3.87GLYY520 pKa = 11.14GEE522 pKa = 4.45TAPFNYY528 pKa = 8.66TRR530 pKa = 11.84WVTEE534 pKa = 3.7EE535 pKa = 3.62NNEE538 pKa = 3.9YY539 pKa = 9.2MEE541 pKa = 4.44QMLSSEE547 pKa = 4.4GFDD550 pKa = 3.6TEE552 pKa = 3.53WRR554 pKa = 11.84AEE556 pKa = 4.18VFNEE560 pKa = 3.42WQEE563 pKa = 4.2YY564 pKa = 9.52FMEE567 pKa = 4.71EE568 pKa = 4.2LPIIPTLFRR577 pKa = 11.84TEE579 pKa = 4.04VFPVNDD585 pKa = 3.27RR586 pKa = 11.84VGAFDD591 pKa = 3.2ITVGVDD597 pKa = 3.15PEE599 pKa = 4.31TAGAEE604 pKa = 4.29TGYY607 pKa = 8.56HH608 pKa = 4.1TWYY611 pKa = 8.88LTSEE615 pKa = 4.18EE616 pKa = 4.72RR617 pKa = 11.84VTEE620 pKa = 4.11
Molecular weight: 68.89 kDa
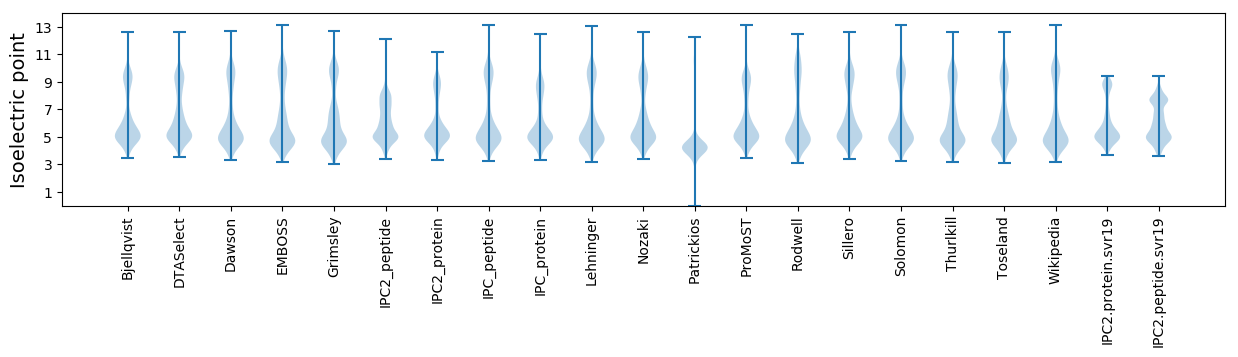
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L4BWW1|A0A1L4BWW1_9LACT DNA starvation/stationary phase protection protein OS=Marinilactibacillus sp. 15R OX=1911586 GN=BKP56_03040 PE=3 SV=1
MM1 pKa = 7.12SQQGLTYY8 pKa = 10.09QPKK11 pKa = 8.71KK12 pKa = 10.19RR13 pKa = 11.84KK14 pKa = 7.43RR15 pKa = 11.84QKK17 pKa = 8.87VHH19 pKa = 5.8GFRR22 pKa = 11.84KK23 pKa = 10.04RR24 pKa = 11.84MSTKK28 pKa = 9.72NGRR31 pKa = 11.84RR32 pKa = 11.84VLKK35 pKa = 10.17ARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 8.51GRR42 pKa = 11.84KK43 pKa = 9.2RR44 pKa = 11.84ISAA47 pKa = 3.72
MM1 pKa = 7.12SQQGLTYY8 pKa = 10.09QPKK11 pKa = 8.71KK12 pKa = 10.19RR13 pKa = 11.84KK14 pKa = 7.43RR15 pKa = 11.84QKK17 pKa = 8.87VHH19 pKa = 5.8GFRR22 pKa = 11.84KK23 pKa = 10.04RR24 pKa = 11.84MSTKK28 pKa = 9.72NGRR31 pKa = 11.84RR32 pKa = 11.84VLKK35 pKa = 10.17ARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 8.51GRR42 pKa = 11.84KK43 pKa = 9.2RR44 pKa = 11.84ISAA47 pKa = 3.72
Molecular weight: 5.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
740988 |
37 |
1529 |
298.1 |
33.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.532 ± 0.048 | 0.505 ± 0.012 |
5.532 ± 0.041 | 8.086 ± 0.057 |
4.34 ± 0.035 | 6.454 ± 0.048 |
1.846 ± 0.022 | 8.087 ± 0.05 |
7.064 ± 0.054 | 9.606 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.654 ± 0.021 | 5.008 ± 0.032 |
3.245 ± 0.025 | 3.793 ± 0.035 |
3.891 ± 0.032 | 6.405 ± 0.036 |
5.667 ± 0.037 | 6.666 ± 0.04 |
0.884 ± 0.017 | 3.736 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
