
Moraxella caviae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae; Moraxella
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
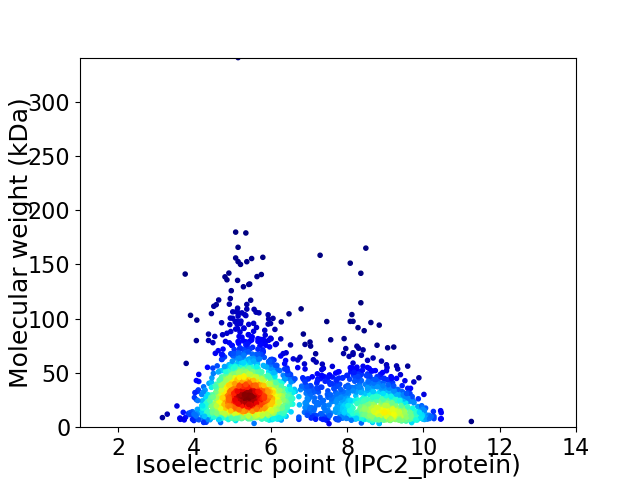
Virtual 2D-PAGE plot for 2269 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S9ZW63|A0A1S9ZW63_9GAMM Sporulation inhibitor kipI OS=Moraxella caviae OX=34060 GN=kipI PE=4 SV=1
MM1 pKa = 7.16SRR3 pKa = 11.84EE4 pKa = 3.78IEE6 pKa = 4.12FEE8 pKa = 3.74ITTDD12 pKa = 3.74SGNLYY17 pKa = 10.59LDD19 pKa = 4.74LEE21 pKa = 4.69GFDD24 pKa = 5.25ANANYY29 pKa = 10.16HH30 pKa = 5.28YY31 pKa = 9.19TLEE34 pKa = 4.48GPISISGAKK43 pKa = 9.34NLPVDD48 pKa = 3.8DD49 pKa = 5.04RR50 pKa = 11.84LFEE53 pKa = 4.25ALYY56 pKa = 10.76DD57 pKa = 4.12DD58 pKa = 4.94EE59 pKa = 6.29LPAGKK64 pKa = 10.28YY65 pKa = 9.3KK66 pKa = 11.12LNIIGNFTDD75 pKa = 3.19IDD77 pKa = 3.6AAVILEE83 pKa = 5.23HH84 pKa = 6.13ITKK87 pKa = 9.2TPAEE91 pKa = 4.44GFTGFDD97 pKa = 3.3APTGNILADD106 pKa = 3.53NTGKK110 pKa = 8.21ITKK113 pKa = 10.15LDD115 pKa = 3.86DD116 pKa = 3.61YY117 pKa = 10.74KK118 pKa = 11.69VKK120 pKa = 11.03VGDD123 pKa = 3.62ATVDD127 pKa = 3.94FSEE130 pKa = 4.6TDD132 pKa = 3.31SHH134 pKa = 7.81SLNLDD139 pKa = 3.3KK140 pKa = 11.2GVLTINKK147 pKa = 9.01DD148 pKa = 2.99GSYY151 pKa = 9.46TYY153 pKa = 10.39QIYY156 pKa = 8.81GHH158 pKa = 6.29NYY160 pKa = 8.85EE161 pKa = 4.79YY162 pKa = 11.27VPSADD167 pKa = 2.85IATTLKK173 pKa = 10.53IQVLDD178 pKa = 4.08AKK180 pKa = 11.31DD181 pKa = 3.81NVLKK185 pKa = 9.61THH187 pKa = 6.6TLNISGDD194 pKa = 3.96TTPPAAGTLTLKK206 pKa = 10.77NFVDD210 pKa = 4.04TGVSDD215 pKa = 4.07SDD217 pKa = 4.0KK218 pKa = 10.25ITNNTYY224 pKa = 7.65FTPVVQDD231 pKa = 3.62AEE233 pKa = 4.18KK234 pKa = 10.06HH235 pKa = 5.03ARR237 pKa = 11.84TIIEE241 pKa = 3.78YY242 pKa = 9.99SANNGEE248 pKa = 4.2YY249 pKa = 9.38WGEE252 pKa = 3.7QRR254 pKa = 11.84EE255 pKa = 4.4EE256 pKa = 3.81NGYY259 pKa = 10.77LNPLQYY265 pKa = 11.1AGDD268 pKa = 3.59GTYY271 pKa = 10.24QIRR274 pKa = 11.84GKK276 pKa = 10.98VIDD279 pKa = 3.61AAGNVSYY286 pKa = 11.1TNVEE290 pKa = 4.19TVTIDD295 pKa = 3.16TTAPTLSEE303 pKa = 4.08QAVSTDD309 pKa = 3.83GIITGKK315 pKa = 8.4TEE317 pKa = 3.79AGATVVAKK325 pKa = 10.58IGDD328 pKa = 4.04EE329 pKa = 4.51VIGTTTADD337 pKa = 3.28KK338 pKa = 11.1DD339 pKa = 4.25GNFSIATGKK348 pKa = 8.64TNGEE352 pKa = 4.38SIQLTITDD360 pKa = 3.34IAGNKK365 pKa = 7.3LTNSYY370 pKa = 10.83SEE372 pKa = 4.12YY373 pKa = 10.71DD374 pKa = 3.39DD375 pKa = 3.44YY376 pKa = 11.7GYY378 pKa = 11.25YY379 pKa = 9.77IGQQEE384 pKa = 4.75DD385 pKa = 4.21DD386 pKa = 4.17FVTITVPDD394 pKa = 4.0TTPPAAGTLVLEE406 pKa = 4.49NFVDD410 pKa = 3.95TGVSDD415 pKa = 3.83SDD417 pKa = 4.24KK418 pKa = 10.43ITDD421 pKa = 4.05NIYY424 pKa = 7.96FTPVVQGAEE433 pKa = 3.72AHH435 pKa = 5.69ATTEE439 pKa = 3.88IEE441 pKa = 4.13YY442 pKa = 10.98SADD445 pKa = 3.52GEE447 pKa = 4.29NWGADD452 pKa = 3.42GVSGDD457 pKa = 3.13GTYY460 pKa = 10.35QIRR463 pKa = 11.84GKK465 pKa = 9.1VTDD468 pKa = 3.32VAGNVSYY475 pKa = 11.21TNVEE479 pKa = 4.19TVTIDD484 pKa = 3.16TTAPTLSEE492 pKa = 4.08QAVSTDD498 pKa = 3.83GIITGKK504 pKa = 8.4TEE506 pKa = 3.83AGATVIATHH515 pKa = 6.23NGEE518 pKa = 4.39KK519 pKa = 10.35VGEE522 pKa = 4.1AVADD526 pKa = 3.89EE527 pKa = 4.64NGNFSIATGKK537 pKa = 6.01TTNNGEE543 pKa = 4.12RR544 pKa = 11.84FEE546 pKa = 5.09LAITDD551 pKa = 3.46IAGNKK556 pKa = 7.24LTEE559 pKa = 3.79RR560 pKa = 11.84HH561 pKa = 5.7GGYY564 pKa = 10.49GEE566 pKa = 3.87YY567 pKa = 10.37DD568 pKa = 3.26EE569 pKa = 4.76YY570 pKa = 11.5GQYY573 pKa = 9.39IWYY576 pKa = 9.62EE577 pKa = 4.27GYY579 pKa = 10.26EE580 pKa = 3.9NDD582 pKa = 4.67YY583 pKa = 9.56ITVIAPDD590 pKa = 4.03TTPPAAGTLALEE602 pKa = 4.78GFTDD606 pKa = 3.8TGVSDD611 pKa = 3.42SDD613 pKa = 4.67KK614 pKa = 8.59ITKK617 pKa = 8.29YY618 pKa = 9.44TFTPVVKK625 pKa = 10.3DD626 pKa = 3.94AEE628 pKa = 4.47AGAYY632 pKa = 8.77TSIEE636 pKa = 4.06YY637 pKa = 8.72STDD640 pKa = 3.15GGEE643 pKa = 4.16TWKK646 pKa = 10.22QWTLTPSQVNDD657 pKa = 2.7HH658 pKa = 5.92WHH660 pKa = 6.67PEE662 pKa = 4.09EE663 pKa = 4.04EE664 pKa = 4.44LTWYY668 pKa = 7.71DD669 pKa = 3.77TPLVSYY675 pKa = 9.84NYY677 pKa = 10.72SSFLSMGIIGRR688 pKa = 11.84TNDD691 pKa = 2.59GTYY694 pKa = 10.18QIRR697 pKa = 11.84GKK699 pKa = 9.2VTDD702 pKa = 3.38TAGNVSYY709 pKa = 10.99TNIEE713 pKa = 4.42TVTLDD718 pKa = 3.2TTAPTISEE726 pKa = 4.13QAISTDD732 pKa = 3.56GVITGITEE740 pKa = 4.25AGATVVAMYY749 pKa = 10.46NGEE752 pKa = 4.27KK753 pKa = 10.27VGEE756 pKa = 4.52AIADD760 pKa = 3.66EE761 pKa = 4.55NGNFSIATGKK771 pKa = 7.72TVNNGEE777 pKa = 4.15RR778 pKa = 11.84FEE780 pKa = 5.05LAITDD785 pKa = 3.41IAGNKK790 pKa = 10.05LEE792 pKa = 4.14VATLVLQDD800 pKa = 3.73FTDD803 pKa = 3.83TGVSDD808 pKa = 3.89SDD810 pKa = 5.22KK811 pKa = 10.26ITNDD815 pKa = 2.9NSFTPAVQDD824 pKa = 3.32AEE826 pKa = 4.31AYY828 pKa = 9.59IMTEE832 pKa = 3.55IEE834 pKa = 4.09YY835 pKa = 8.69STDD838 pKa = 4.68GGNTWQSSNNSYY850 pKa = 10.12WSDD853 pKa = 2.52GTYY856 pKa = 10.21QIRR859 pKa = 11.84GKK861 pKa = 10.27VADD864 pKa = 3.95YY865 pKa = 11.24NDD867 pKa = 3.77DD868 pKa = 3.3VSYY871 pKa = 11.33TNVEE875 pKa = 4.36TVTVDD880 pKa = 3.11TTPPEE885 pKa = 3.96FTYY888 pKa = 10.86TEE890 pKa = 4.45LLDD893 pKa = 4.67DD894 pKa = 4.24NQTITGITEE903 pKa = 4.22AFATVTADD911 pKa = 2.97NGTSATANEE920 pKa = 4.38NGSFSIDD927 pKa = 2.9LGAPYY932 pKa = 10.05TSGEE936 pKa = 4.23MIGFSAMDD944 pKa = 3.68LAGNYY949 pKa = 9.26SHH951 pKa = 7.93DD952 pKa = 4.02YY953 pKa = 10.37VYY955 pKa = 11.26APIMM959 pKa = 4.07
MM1 pKa = 7.16SRR3 pKa = 11.84EE4 pKa = 3.78IEE6 pKa = 4.12FEE8 pKa = 3.74ITTDD12 pKa = 3.74SGNLYY17 pKa = 10.59LDD19 pKa = 4.74LEE21 pKa = 4.69GFDD24 pKa = 5.25ANANYY29 pKa = 10.16HH30 pKa = 5.28YY31 pKa = 9.19TLEE34 pKa = 4.48GPISISGAKK43 pKa = 9.34NLPVDD48 pKa = 3.8DD49 pKa = 5.04RR50 pKa = 11.84LFEE53 pKa = 4.25ALYY56 pKa = 10.76DD57 pKa = 4.12DD58 pKa = 4.94EE59 pKa = 6.29LPAGKK64 pKa = 10.28YY65 pKa = 9.3KK66 pKa = 11.12LNIIGNFTDD75 pKa = 3.19IDD77 pKa = 3.6AAVILEE83 pKa = 5.23HH84 pKa = 6.13ITKK87 pKa = 9.2TPAEE91 pKa = 4.44GFTGFDD97 pKa = 3.3APTGNILADD106 pKa = 3.53NTGKK110 pKa = 8.21ITKK113 pKa = 10.15LDD115 pKa = 3.86DD116 pKa = 3.61YY117 pKa = 10.74KK118 pKa = 11.69VKK120 pKa = 11.03VGDD123 pKa = 3.62ATVDD127 pKa = 3.94FSEE130 pKa = 4.6TDD132 pKa = 3.31SHH134 pKa = 7.81SLNLDD139 pKa = 3.3KK140 pKa = 11.2GVLTINKK147 pKa = 9.01DD148 pKa = 2.99GSYY151 pKa = 9.46TYY153 pKa = 10.39QIYY156 pKa = 8.81GHH158 pKa = 6.29NYY160 pKa = 8.85EE161 pKa = 4.79YY162 pKa = 11.27VPSADD167 pKa = 2.85IATTLKK173 pKa = 10.53IQVLDD178 pKa = 4.08AKK180 pKa = 11.31DD181 pKa = 3.81NVLKK185 pKa = 9.61THH187 pKa = 6.6TLNISGDD194 pKa = 3.96TTPPAAGTLTLKK206 pKa = 10.77NFVDD210 pKa = 4.04TGVSDD215 pKa = 4.07SDD217 pKa = 4.0KK218 pKa = 10.25ITNNTYY224 pKa = 7.65FTPVVQDD231 pKa = 3.62AEE233 pKa = 4.18KK234 pKa = 10.06HH235 pKa = 5.03ARR237 pKa = 11.84TIIEE241 pKa = 3.78YY242 pKa = 9.99SANNGEE248 pKa = 4.2YY249 pKa = 9.38WGEE252 pKa = 3.7QRR254 pKa = 11.84EE255 pKa = 4.4EE256 pKa = 3.81NGYY259 pKa = 10.77LNPLQYY265 pKa = 11.1AGDD268 pKa = 3.59GTYY271 pKa = 10.24QIRR274 pKa = 11.84GKK276 pKa = 10.98VIDD279 pKa = 3.61AAGNVSYY286 pKa = 11.1TNVEE290 pKa = 4.19TVTIDD295 pKa = 3.16TTAPTLSEE303 pKa = 4.08QAVSTDD309 pKa = 3.83GIITGKK315 pKa = 8.4TEE317 pKa = 3.79AGATVVAKK325 pKa = 10.58IGDD328 pKa = 4.04EE329 pKa = 4.51VIGTTTADD337 pKa = 3.28KK338 pKa = 11.1DD339 pKa = 4.25GNFSIATGKK348 pKa = 8.64TNGEE352 pKa = 4.38SIQLTITDD360 pKa = 3.34IAGNKK365 pKa = 7.3LTNSYY370 pKa = 10.83SEE372 pKa = 4.12YY373 pKa = 10.71DD374 pKa = 3.39DD375 pKa = 3.44YY376 pKa = 11.7GYY378 pKa = 11.25YY379 pKa = 9.77IGQQEE384 pKa = 4.75DD385 pKa = 4.21DD386 pKa = 4.17FVTITVPDD394 pKa = 4.0TTPPAAGTLVLEE406 pKa = 4.49NFVDD410 pKa = 3.95TGVSDD415 pKa = 3.83SDD417 pKa = 4.24KK418 pKa = 10.43ITDD421 pKa = 4.05NIYY424 pKa = 7.96FTPVVQGAEE433 pKa = 3.72AHH435 pKa = 5.69ATTEE439 pKa = 3.88IEE441 pKa = 4.13YY442 pKa = 10.98SADD445 pKa = 3.52GEE447 pKa = 4.29NWGADD452 pKa = 3.42GVSGDD457 pKa = 3.13GTYY460 pKa = 10.35QIRR463 pKa = 11.84GKK465 pKa = 9.1VTDD468 pKa = 3.32VAGNVSYY475 pKa = 11.21TNVEE479 pKa = 4.19TVTIDD484 pKa = 3.16TTAPTLSEE492 pKa = 4.08QAVSTDD498 pKa = 3.83GIITGKK504 pKa = 8.4TEE506 pKa = 3.83AGATVIATHH515 pKa = 6.23NGEE518 pKa = 4.39KK519 pKa = 10.35VGEE522 pKa = 4.1AVADD526 pKa = 3.89EE527 pKa = 4.64NGNFSIATGKK537 pKa = 6.01TTNNGEE543 pKa = 4.12RR544 pKa = 11.84FEE546 pKa = 5.09LAITDD551 pKa = 3.46IAGNKK556 pKa = 7.24LTEE559 pKa = 3.79RR560 pKa = 11.84HH561 pKa = 5.7GGYY564 pKa = 10.49GEE566 pKa = 3.87YY567 pKa = 10.37DD568 pKa = 3.26EE569 pKa = 4.76YY570 pKa = 11.5GQYY573 pKa = 9.39IWYY576 pKa = 9.62EE577 pKa = 4.27GYY579 pKa = 10.26EE580 pKa = 3.9NDD582 pKa = 4.67YY583 pKa = 9.56ITVIAPDD590 pKa = 4.03TTPPAAGTLALEE602 pKa = 4.78GFTDD606 pKa = 3.8TGVSDD611 pKa = 3.42SDD613 pKa = 4.67KK614 pKa = 8.59ITKK617 pKa = 8.29YY618 pKa = 9.44TFTPVVKK625 pKa = 10.3DD626 pKa = 3.94AEE628 pKa = 4.47AGAYY632 pKa = 8.77TSIEE636 pKa = 4.06YY637 pKa = 8.72STDD640 pKa = 3.15GGEE643 pKa = 4.16TWKK646 pKa = 10.22QWTLTPSQVNDD657 pKa = 2.7HH658 pKa = 5.92WHH660 pKa = 6.67PEE662 pKa = 4.09EE663 pKa = 4.04EE664 pKa = 4.44LTWYY668 pKa = 7.71DD669 pKa = 3.77TPLVSYY675 pKa = 9.84NYY677 pKa = 10.72SSFLSMGIIGRR688 pKa = 11.84TNDD691 pKa = 2.59GTYY694 pKa = 10.18QIRR697 pKa = 11.84GKK699 pKa = 9.2VTDD702 pKa = 3.38TAGNVSYY709 pKa = 10.99TNIEE713 pKa = 4.42TVTLDD718 pKa = 3.2TTAPTISEE726 pKa = 4.13QAISTDD732 pKa = 3.56GVITGITEE740 pKa = 4.25AGATVVAMYY749 pKa = 10.46NGEE752 pKa = 4.27KK753 pKa = 10.27VGEE756 pKa = 4.52AIADD760 pKa = 3.66EE761 pKa = 4.55NGNFSIATGKK771 pKa = 7.72TVNNGEE777 pKa = 4.15RR778 pKa = 11.84FEE780 pKa = 5.05LAITDD785 pKa = 3.41IAGNKK790 pKa = 10.05LEE792 pKa = 4.14VATLVLQDD800 pKa = 3.73FTDD803 pKa = 3.83TGVSDD808 pKa = 3.89SDD810 pKa = 5.22KK811 pKa = 10.26ITNDD815 pKa = 2.9NSFTPAVQDD824 pKa = 3.32AEE826 pKa = 4.31AYY828 pKa = 9.59IMTEE832 pKa = 3.55IEE834 pKa = 4.09YY835 pKa = 8.69STDD838 pKa = 4.68GGNTWQSSNNSYY850 pKa = 10.12WSDD853 pKa = 2.52GTYY856 pKa = 10.21QIRR859 pKa = 11.84GKK861 pKa = 10.27VADD864 pKa = 3.95YY865 pKa = 11.24NDD867 pKa = 3.77DD868 pKa = 3.3VSYY871 pKa = 11.33TNVEE875 pKa = 4.36TVTVDD880 pKa = 3.11TTPPEE885 pKa = 3.96FTYY888 pKa = 10.86TEE890 pKa = 4.45LLDD893 pKa = 4.67DD894 pKa = 4.24NQTITGITEE903 pKa = 4.22AFATVTADD911 pKa = 2.97NGTSATANEE920 pKa = 4.38NGSFSIDD927 pKa = 2.9LGAPYY932 pKa = 10.05TSGEE936 pKa = 4.23MIGFSAMDD944 pKa = 3.68LAGNYY949 pKa = 9.26SHH951 pKa = 7.93DD952 pKa = 4.02YY953 pKa = 10.37VYY955 pKa = 11.26APIMM959 pKa = 4.07
Molecular weight: 103.06 kDa
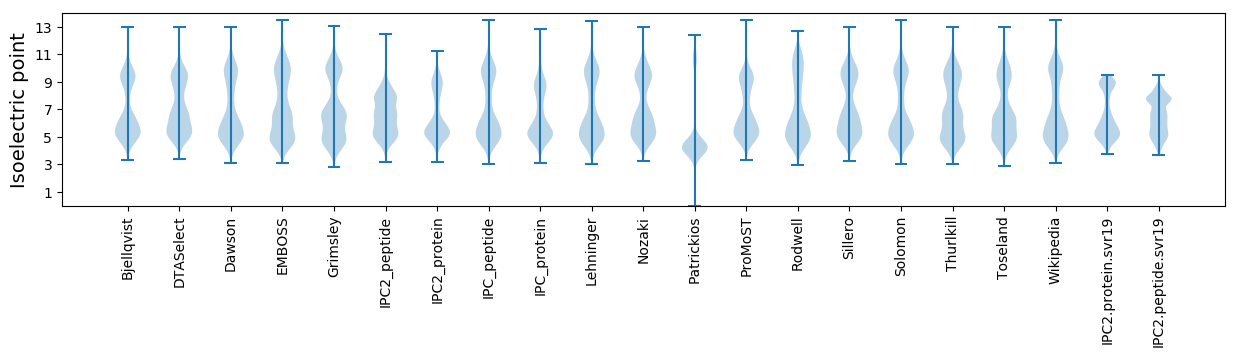
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S9ZXP2|A0A1S9ZXP2_9GAMM Domain of uncharacterized function (DUF477) OS=Moraxella caviae OX=34060 GN=B0181_09810 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.32KK26 pKa = 10.31GRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.03GRR39 pKa = 11.84HH40 pKa = 5.34RR41 pKa = 11.84LTVV44 pKa = 3.07
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.32KK26 pKa = 10.31GRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.03GRR39 pKa = 11.84HH40 pKa = 5.34RR41 pKa = 11.84LTVV44 pKa = 3.07
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
672763 |
26 |
3231 |
296.5 |
32.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.705 ± 0.081 | 0.977 ± 0.018 |
5.987 ± 0.043 | 5.247 ± 0.064 |
4.073 ± 0.041 | 6.982 ± 0.053 |
2.418 ± 0.028 | 5.942 ± 0.042 |
5.486 ± 0.049 | 10.052 ± 0.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.54 ± 0.029 | 4.316 ± 0.05 |
3.856 ± 0.033 | 4.328 ± 0.044 |
4.506 ± 0.046 | 5.778 ± 0.044 |
5.741 ± 0.057 | 7.018 ± 0.05 |
1.142 ± 0.02 | 2.906 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
