
Confluentibacter flavum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Confluentibacter
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
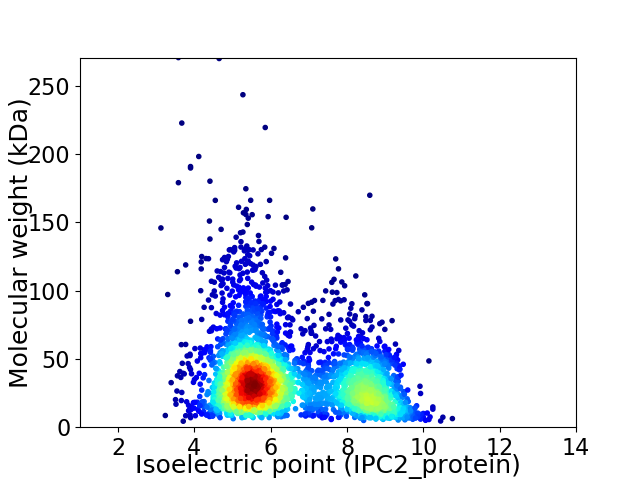
Virtual 2D-PAGE plot for 3459 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N3HLW7|A0A2N3HLW7_9FLAO Cysteine synthase OS=Confluentibacter flavum OX=1909700 GN=cysK PE=3 SV=1
MM1 pKa = 7.24VPQIKK6 pKa = 9.91IFCNFIYY13 pKa = 9.99FIFILLSYY21 pKa = 10.73NAFAQSWSQSILTFSGTGNVNGGVTSLMYY50 pKa = 10.96GPDD53 pKa = 2.99NRR55 pKa = 11.84LYY57 pKa = 9.25VAEE60 pKa = 4.01YY61 pKa = 9.29TGAIKK66 pKa = 10.53ILTIQRR72 pKa = 11.84NSPINYY78 pKa = 8.16VATNVEE84 pKa = 4.03ILDD87 pKa = 5.04GIRR90 pKa = 11.84TMPDD94 pKa = 2.91HH95 pKa = 7.14NDD97 pKa = 3.28DD98 pKa = 3.64GTLYY102 pKa = 10.91SSLNRR107 pKa = 11.84EE108 pKa = 4.28TLGLTVVGTPTNPVIYY124 pKa = 9.21VTSSDD129 pKa = 3.48FRR131 pKa = 11.84IGAGEE136 pKa = 4.04GGGNGDD142 pKa = 4.1VNLDD146 pKa = 3.77TNSGVITRR154 pKa = 11.84FSWNGSAWNVVDD166 pKa = 4.41IVRR169 pKa = 11.84GLPRR173 pKa = 11.84SEE175 pKa = 4.39EE176 pKa = 3.77NHH178 pKa = 5.53ATNGLEE184 pKa = 3.74FVTINGTDD192 pKa = 3.73YY193 pKa = 11.43LIVAQGGHH201 pKa = 5.48TNGGSPSTNFVYY213 pKa = 10.41SCEE216 pKa = 4.12YY217 pKa = 10.32ALSGAILAVNLTALEE232 pKa = 4.18TMPILDD238 pKa = 4.46DD239 pKa = 3.59NGRR242 pKa = 11.84AYY244 pKa = 9.65IYY246 pKa = 9.82DD247 pKa = 4.85LPTLDD252 pKa = 5.59DD253 pKa = 3.58PTRR256 pKa = 11.84PNIHH260 pKa = 7.17ADD262 pKa = 3.57GSPASEE268 pKa = 5.5DD269 pKa = 3.69PDD271 pKa = 3.95DD272 pKa = 4.84PNYY275 pKa = 11.16SPLDD279 pKa = 3.56INDD282 pKa = 3.34PWGGNDD288 pKa = 4.45GLNQAILVPGGPVQILSPGYY308 pKa = 9.46RR309 pKa = 11.84NAYY312 pKa = 9.24DD313 pKa = 3.68LVVTEE318 pKa = 4.98SGALYY323 pKa = 9.26VTDD326 pKa = 3.85NGANGGWGGLPMNEE340 pKa = 4.28GGGSVTNDD348 pKa = 3.32YY349 pKa = 11.34NPAEE353 pKa = 4.71PGSSDD358 pKa = 3.18SLLYY362 pKa = 11.23GEE364 pKa = 4.89MVNNLDD370 pKa = 3.65HH371 pKa = 6.61LQLVTTNLQAYY382 pKa = 9.85AFGTLPEE389 pKa = 4.39TYY391 pKa = 10.22YY392 pKa = 10.95GGHH395 pKa = 7.19PNPTRR400 pKa = 11.84ANPVGAGLYY409 pKa = 8.79TADD412 pKa = 4.63GINNVFRR419 pKa = 11.84TLKK422 pKa = 10.08YY423 pKa = 10.78DD424 pKa = 3.98PDD426 pKa = 4.09GSTPGSTSNPNEE438 pKa = 4.17ALPVNWPPLPVSEE451 pKa = 4.66ANTVEE456 pKa = 4.25GDD458 pKa = 2.92WRR460 pKa = 11.84GPGMINPDD468 pKa = 4.11GPNDD472 pKa = 3.87DD473 pKa = 5.15PVVTWGTNTNGIDD486 pKa = 3.68EE487 pKa = 4.46YY488 pKa = 10.41TASNFGGVMQGNLLAGTHH506 pKa = 6.22NGLIRR511 pKa = 11.84RR512 pKa = 11.84VQLNANGTLASGGLTSNFLSGMGGNALGITCNSDD546 pKa = 3.05FDD548 pKa = 4.22IFPGTVWAGTLNGKK562 pKa = 9.23IVVFEE567 pKa = 3.96PTDD570 pKa = 3.61FLNCLDD576 pKa = 4.85PSDD579 pKa = 4.66PLYY582 pKa = 10.97DD583 pKa = 3.82ANADD587 pKa = 3.79YY588 pKa = 11.32DD589 pKa = 3.92SDD591 pKa = 4.47GYY593 pKa = 11.5SNQDD597 pKa = 3.17EE598 pKa = 4.55EE599 pKa = 5.8DD600 pKa = 3.8NGTEE604 pKa = 3.67ICNGGSQPDD613 pKa = 3.6DD614 pKa = 3.4WDD616 pKa = 3.53KK617 pKa = 12.1VMGGQLISDD626 pKa = 4.74LNDD629 pKa = 4.12PDD631 pKa = 5.81DD632 pKa = 5.53DD633 pKa = 4.91FDD635 pKa = 5.96GIPDD639 pKa = 4.01ALDD642 pKa = 3.78PFQLGDD648 pKa = 3.64PTVEE652 pKa = 4.03GSDD655 pKa = 3.35AFTIPILNDD664 pKa = 3.51FFNDD668 pKa = 3.48QQNLGGIYY676 pKa = 10.53GLGLTGLMNNGDD688 pKa = 3.8TGANWLLWLDD698 pKa = 4.53RR699 pKa = 11.84KK700 pKa = 10.6DD701 pKa = 5.34DD702 pKa = 4.61PNDD705 pKa = 3.89PNPNDD710 pKa = 3.76ILGGAPGLMTSQMTSGTALGTSNNQEE736 pKa = 3.44KK737 pKa = 10.32GYY739 pKa = 10.56QYY741 pKa = 11.22GVQIQNDD748 pKa = 3.62SGIITVSGGMVGFNGPLTLYY768 pKa = 9.82EE769 pKa = 4.29NLSAPSGEE777 pKa = 4.04LGFFIGDD784 pKa = 3.93GTQSNYY790 pKa = 10.1IKK792 pKa = 10.61FVVTTTGFVALQEE805 pKa = 4.52IEE807 pKa = 5.82DD808 pKa = 4.28IPQSPITYY816 pKa = 9.82NLPQANRR823 pKa = 11.84PTSGIKK829 pKa = 10.12FFFEE833 pKa = 4.04IDD835 pKa = 3.48PSIGQVTLKK844 pKa = 10.51FQIDD848 pKa = 3.88EE849 pKa = 4.6GSLTTLGTIVAQGSILQALQSSSTDD874 pKa = 3.46LAVGLIGASNEE885 pKa = 4.12DD886 pKa = 3.65GVEE889 pKa = 5.15LEE891 pKa = 4.44GTWDD895 pKa = 3.77FLNVTAEE902 pKa = 4.1KK903 pKa = 10.25PYY905 pKa = 9.99MVQNIPNLQRR915 pKa = 11.84EE916 pKa = 4.43SGAPDD921 pKa = 4.11DD922 pKa = 5.41MFDD925 pKa = 4.38LDD927 pKa = 6.57DD928 pKa = 4.36YY929 pKa = 11.91FEE931 pKa = 5.82DD932 pKa = 4.24NNGTANLTYY941 pKa = 10.67SVALNTNPNILASISNNLLTITYY964 pKa = 9.31PSEE967 pKa = 3.89LQTADD972 pKa = 2.82ITIRR976 pKa = 11.84ASDD979 pKa = 3.61LNGYY983 pKa = 8.27FVEE986 pKa = 4.55DD987 pKa = 3.93TFTVSIINSSLILARR1002 pKa = 11.84VNAGGAAVSATDD1014 pKa = 5.01GGANWEE1020 pKa = 4.24QNNVAGATSGPSYY1033 pKa = 10.95SVNTGTIYY1041 pKa = 10.96NGGLLNINRR1050 pKa = 11.84HH1051 pKa = 4.87SSIPDD1056 pKa = 3.62YY1057 pKa = 10.86IDD1059 pKa = 3.22EE1060 pKa = 4.32STYY1063 pKa = 11.16NAIFAYY1069 pKa = 9.71EE1070 pKa = 4.25RR1071 pKa = 11.84YY1072 pKa = 9.89DD1073 pKa = 3.43VTASPEE1079 pKa = 3.9MEE1081 pKa = 4.09FTFVLQNGDD1090 pKa = 3.83YY1091 pKa = 10.37QVNLYY1096 pKa = 8.86MGNSFVGTSEE1106 pKa = 3.41IGDD1109 pKa = 3.47RR1110 pKa = 11.84VFDD1113 pKa = 3.73IEE1115 pKa = 4.18IEE1117 pKa = 4.28GVIVEE1122 pKa = 4.97DD1123 pKa = 4.58NMDD1126 pKa = 4.84LIARR1130 pKa = 11.84FGHH1133 pKa = 5.6QSGGMVTYY1141 pKa = 9.79PVTVSGGVLNIQFLHH1156 pKa = 5.79EE1157 pKa = 4.36VEE1159 pKa = 4.75NPLLNAIEE1167 pKa = 4.36IMAAPPQYY1175 pKa = 10.27PPLSVNPIANQTNLTGDD1192 pKa = 3.86VISTLGVSASGGNPNNNFTYY1212 pKa = 10.55SIDD1215 pKa = 3.8GQPLGINIEE1224 pKa = 4.16PTNGLIYY1231 pKa = 9.34GTIDD1235 pKa = 3.11QTASNGGTNNDD1246 pKa = 3.28GVHH1249 pKa = 5.87GVTVTVEE1256 pKa = 4.43KK1257 pKa = 10.91VGSTSVTTNFSWTVNDD1273 pKa = 5.07LAWFDD1278 pKa = 3.87KK1279 pKa = 11.33NEE1281 pKa = 4.03NEE1283 pKa = 4.5NYY1285 pKa = 7.42TARR1288 pKa = 11.84HH1289 pKa = 5.42EE1290 pKa = 4.43CSFVQAGDD1298 pKa = 3.21KK1299 pKa = 10.58FYY1301 pKa = 11.44LLGGRR1306 pKa = 11.84EE1307 pKa = 3.93NSKK1310 pKa = 8.31TLDD1313 pKa = 3.41VYY1315 pKa = 11.5NYY1317 pKa = 9.34TSNSWNSLVDD1327 pKa = 3.78SAPMEE1332 pKa = 4.47FNHH1335 pKa = 6.4FQATEE1340 pKa = 3.94YY1341 pKa = 10.59QGLIWVIGAFQTNSFPNEE1359 pKa = 3.82TPADD1363 pKa = 4.47FIWAFDD1369 pKa = 3.69PVAQQWIQGPEE1380 pKa = 3.73IPSGRR1385 pKa = 11.84RR1386 pKa = 11.84RR1387 pKa = 11.84GSAGLVVHH1395 pKa = 6.63NDD1397 pKa = 2.33KK1398 pKa = 11.02FYY1400 pKa = 10.56IVGGNTIGHH1409 pKa = 6.3NGGYY1413 pKa = 9.77VPWFDD1418 pKa = 4.53EE1419 pKa = 4.25YY1420 pKa = 11.74DD1421 pKa = 3.68PVLGTWTTLTNAPRR1435 pKa = 11.84SRR1437 pKa = 11.84DD1438 pKa = 3.32HH1439 pKa = 5.98FHH1441 pKa = 7.21AVVINNKK1448 pKa = 9.77LYY1450 pKa = 10.3AASGRR1455 pKa = 11.84LSGGDD1460 pKa = 3.13GGTFAPVIPEE1470 pKa = 3.52VDD1472 pKa = 3.49VYY1474 pKa = 11.66DD1475 pKa = 4.8FSTNSWSSLPSDD1487 pKa = 3.72QNIPTPRR1494 pKa = 11.84AGAIVANFNNKK1505 pKa = 8.84LVIAGGEE1512 pKa = 3.96VSTNVNALDD1521 pKa = 3.14ITEE1524 pKa = 4.94IYY1526 pKa = 10.52DD1527 pKa = 4.14PLDD1530 pKa = 3.57QSWSTGASLIHH1541 pKa = 6.57PRR1543 pKa = 11.84HH1544 pKa = 5.0GTQGIVSGNGIFVVAGSPVMGGGKK1568 pKa = 8.8QKK1570 pKa = 10.94NMEE1573 pKa = 4.52FYY1575 pKa = 10.98GSDD1578 pKa = 3.64SPTGIPLDD1586 pKa = 3.76ASEE1589 pKa = 5.27LMGPTTVDD1597 pKa = 3.7FSVQNSQDD1605 pKa = 2.85ITLDD1609 pKa = 3.67LEE1611 pKa = 4.49NGNTGIFIRR1620 pKa = 11.84SVEE1623 pKa = 4.24INGHH1627 pKa = 6.6DD1628 pKa = 4.32DD1629 pKa = 3.74DD1630 pKa = 6.52DD1631 pKa = 4.98FVIQSGNLNNVLLKK1645 pKa = 10.9SNSSKK1650 pKa = 10.51IISVDD1655 pKa = 4.11FIGAGTEE1662 pKa = 3.88KK1663 pKa = 10.82NAILTIDD1670 pKa = 3.52YY1671 pKa = 8.84GANASLDD1678 pKa = 3.15ISLSYY1683 pKa = 11.19DD1684 pKa = 3.46STLSSPDD1691 pKa = 3.17EE1692 pKa = 4.34KK1693 pKa = 11.61DD1694 pKa = 3.34NISNVFTIYY1703 pKa = 10.36PNPSSEE1709 pKa = 4.05YY1710 pKa = 8.91VYY1712 pKa = 11.31VEE1714 pKa = 4.34IKK1716 pKa = 10.56SSSIDD1721 pKa = 3.27VTDD1724 pKa = 4.65FLLFDD1729 pKa = 3.15ITGRR1733 pKa = 11.84LIKK1736 pKa = 10.15KK1737 pKa = 9.48IKK1739 pKa = 10.04NDD1741 pKa = 3.16NSEE1744 pKa = 3.51QSTFKK1749 pKa = 10.83KK1750 pKa = 10.58QIDD1753 pKa = 3.69VRR1755 pKa = 11.84GLQSGLYY1762 pKa = 8.59TLYY1765 pKa = 10.17ILSKK1769 pKa = 10.52GSVVHH1774 pKa = 6.88KK1775 pKa = 10.7ARR1777 pKa = 11.84LVINNN1782 pKa = 3.78
MM1 pKa = 7.24VPQIKK6 pKa = 9.91IFCNFIYY13 pKa = 9.99FIFILLSYY21 pKa = 10.73NAFAQSWSQSILTFSGTGNVNGGVTSLMYY50 pKa = 10.96GPDD53 pKa = 2.99NRR55 pKa = 11.84LYY57 pKa = 9.25VAEE60 pKa = 4.01YY61 pKa = 9.29TGAIKK66 pKa = 10.53ILTIQRR72 pKa = 11.84NSPINYY78 pKa = 8.16VATNVEE84 pKa = 4.03ILDD87 pKa = 5.04GIRR90 pKa = 11.84TMPDD94 pKa = 2.91HH95 pKa = 7.14NDD97 pKa = 3.28DD98 pKa = 3.64GTLYY102 pKa = 10.91SSLNRR107 pKa = 11.84EE108 pKa = 4.28TLGLTVVGTPTNPVIYY124 pKa = 9.21VTSSDD129 pKa = 3.48FRR131 pKa = 11.84IGAGEE136 pKa = 4.04GGGNGDD142 pKa = 4.1VNLDD146 pKa = 3.77TNSGVITRR154 pKa = 11.84FSWNGSAWNVVDD166 pKa = 4.41IVRR169 pKa = 11.84GLPRR173 pKa = 11.84SEE175 pKa = 4.39EE176 pKa = 3.77NHH178 pKa = 5.53ATNGLEE184 pKa = 3.74FVTINGTDD192 pKa = 3.73YY193 pKa = 11.43LIVAQGGHH201 pKa = 5.48TNGGSPSTNFVYY213 pKa = 10.41SCEE216 pKa = 4.12YY217 pKa = 10.32ALSGAILAVNLTALEE232 pKa = 4.18TMPILDD238 pKa = 4.46DD239 pKa = 3.59NGRR242 pKa = 11.84AYY244 pKa = 9.65IYY246 pKa = 9.82DD247 pKa = 4.85LPTLDD252 pKa = 5.59DD253 pKa = 3.58PTRR256 pKa = 11.84PNIHH260 pKa = 7.17ADD262 pKa = 3.57GSPASEE268 pKa = 5.5DD269 pKa = 3.69PDD271 pKa = 3.95DD272 pKa = 4.84PNYY275 pKa = 11.16SPLDD279 pKa = 3.56INDD282 pKa = 3.34PWGGNDD288 pKa = 4.45GLNQAILVPGGPVQILSPGYY308 pKa = 9.46RR309 pKa = 11.84NAYY312 pKa = 9.24DD313 pKa = 3.68LVVTEE318 pKa = 4.98SGALYY323 pKa = 9.26VTDD326 pKa = 3.85NGANGGWGGLPMNEE340 pKa = 4.28GGGSVTNDD348 pKa = 3.32YY349 pKa = 11.34NPAEE353 pKa = 4.71PGSSDD358 pKa = 3.18SLLYY362 pKa = 11.23GEE364 pKa = 4.89MVNNLDD370 pKa = 3.65HH371 pKa = 6.61LQLVTTNLQAYY382 pKa = 9.85AFGTLPEE389 pKa = 4.39TYY391 pKa = 10.22YY392 pKa = 10.95GGHH395 pKa = 7.19PNPTRR400 pKa = 11.84ANPVGAGLYY409 pKa = 8.79TADD412 pKa = 4.63GINNVFRR419 pKa = 11.84TLKK422 pKa = 10.08YY423 pKa = 10.78DD424 pKa = 3.98PDD426 pKa = 4.09GSTPGSTSNPNEE438 pKa = 4.17ALPVNWPPLPVSEE451 pKa = 4.66ANTVEE456 pKa = 4.25GDD458 pKa = 2.92WRR460 pKa = 11.84GPGMINPDD468 pKa = 4.11GPNDD472 pKa = 3.87DD473 pKa = 5.15PVVTWGTNTNGIDD486 pKa = 3.68EE487 pKa = 4.46YY488 pKa = 10.41TASNFGGVMQGNLLAGTHH506 pKa = 6.22NGLIRR511 pKa = 11.84RR512 pKa = 11.84VQLNANGTLASGGLTSNFLSGMGGNALGITCNSDD546 pKa = 3.05FDD548 pKa = 4.22IFPGTVWAGTLNGKK562 pKa = 9.23IVVFEE567 pKa = 3.96PTDD570 pKa = 3.61FLNCLDD576 pKa = 4.85PSDD579 pKa = 4.66PLYY582 pKa = 10.97DD583 pKa = 3.82ANADD587 pKa = 3.79YY588 pKa = 11.32DD589 pKa = 3.92SDD591 pKa = 4.47GYY593 pKa = 11.5SNQDD597 pKa = 3.17EE598 pKa = 4.55EE599 pKa = 5.8DD600 pKa = 3.8NGTEE604 pKa = 3.67ICNGGSQPDD613 pKa = 3.6DD614 pKa = 3.4WDD616 pKa = 3.53KK617 pKa = 12.1VMGGQLISDD626 pKa = 4.74LNDD629 pKa = 4.12PDD631 pKa = 5.81DD632 pKa = 5.53DD633 pKa = 4.91FDD635 pKa = 5.96GIPDD639 pKa = 4.01ALDD642 pKa = 3.78PFQLGDD648 pKa = 3.64PTVEE652 pKa = 4.03GSDD655 pKa = 3.35AFTIPILNDD664 pKa = 3.51FFNDD668 pKa = 3.48QQNLGGIYY676 pKa = 10.53GLGLTGLMNNGDD688 pKa = 3.8TGANWLLWLDD698 pKa = 4.53RR699 pKa = 11.84KK700 pKa = 10.6DD701 pKa = 5.34DD702 pKa = 4.61PNDD705 pKa = 3.89PNPNDD710 pKa = 3.76ILGGAPGLMTSQMTSGTALGTSNNQEE736 pKa = 3.44KK737 pKa = 10.32GYY739 pKa = 10.56QYY741 pKa = 11.22GVQIQNDD748 pKa = 3.62SGIITVSGGMVGFNGPLTLYY768 pKa = 9.82EE769 pKa = 4.29NLSAPSGEE777 pKa = 4.04LGFFIGDD784 pKa = 3.93GTQSNYY790 pKa = 10.1IKK792 pKa = 10.61FVVTTTGFVALQEE805 pKa = 4.52IEE807 pKa = 5.82DD808 pKa = 4.28IPQSPITYY816 pKa = 9.82NLPQANRR823 pKa = 11.84PTSGIKK829 pKa = 10.12FFFEE833 pKa = 4.04IDD835 pKa = 3.48PSIGQVTLKK844 pKa = 10.51FQIDD848 pKa = 3.88EE849 pKa = 4.6GSLTTLGTIVAQGSILQALQSSSTDD874 pKa = 3.46LAVGLIGASNEE885 pKa = 4.12DD886 pKa = 3.65GVEE889 pKa = 5.15LEE891 pKa = 4.44GTWDD895 pKa = 3.77FLNVTAEE902 pKa = 4.1KK903 pKa = 10.25PYY905 pKa = 9.99MVQNIPNLQRR915 pKa = 11.84EE916 pKa = 4.43SGAPDD921 pKa = 4.11DD922 pKa = 5.41MFDD925 pKa = 4.38LDD927 pKa = 6.57DD928 pKa = 4.36YY929 pKa = 11.91FEE931 pKa = 5.82DD932 pKa = 4.24NNGTANLTYY941 pKa = 10.67SVALNTNPNILASISNNLLTITYY964 pKa = 9.31PSEE967 pKa = 3.89LQTADD972 pKa = 2.82ITIRR976 pKa = 11.84ASDD979 pKa = 3.61LNGYY983 pKa = 8.27FVEE986 pKa = 4.55DD987 pKa = 3.93TFTVSIINSSLILARR1002 pKa = 11.84VNAGGAAVSATDD1014 pKa = 5.01GGANWEE1020 pKa = 4.24QNNVAGATSGPSYY1033 pKa = 10.95SVNTGTIYY1041 pKa = 10.96NGGLLNINRR1050 pKa = 11.84HH1051 pKa = 4.87SSIPDD1056 pKa = 3.62YY1057 pKa = 10.86IDD1059 pKa = 3.22EE1060 pKa = 4.32STYY1063 pKa = 11.16NAIFAYY1069 pKa = 9.71EE1070 pKa = 4.25RR1071 pKa = 11.84YY1072 pKa = 9.89DD1073 pKa = 3.43VTASPEE1079 pKa = 3.9MEE1081 pKa = 4.09FTFVLQNGDD1090 pKa = 3.83YY1091 pKa = 10.37QVNLYY1096 pKa = 8.86MGNSFVGTSEE1106 pKa = 3.41IGDD1109 pKa = 3.47RR1110 pKa = 11.84VFDD1113 pKa = 3.73IEE1115 pKa = 4.18IEE1117 pKa = 4.28GVIVEE1122 pKa = 4.97DD1123 pKa = 4.58NMDD1126 pKa = 4.84LIARR1130 pKa = 11.84FGHH1133 pKa = 5.6QSGGMVTYY1141 pKa = 9.79PVTVSGGVLNIQFLHH1156 pKa = 5.79EE1157 pKa = 4.36VEE1159 pKa = 4.75NPLLNAIEE1167 pKa = 4.36IMAAPPQYY1175 pKa = 10.27PPLSVNPIANQTNLTGDD1192 pKa = 3.86VISTLGVSASGGNPNNNFTYY1212 pKa = 10.55SIDD1215 pKa = 3.8GQPLGINIEE1224 pKa = 4.16PTNGLIYY1231 pKa = 9.34GTIDD1235 pKa = 3.11QTASNGGTNNDD1246 pKa = 3.28GVHH1249 pKa = 5.87GVTVTVEE1256 pKa = 4.43KK1257 pKa = 10.91VGSTSVTTNFSWTVNDD1273 pKa = 5.07LAWFDD1278 pKa = 3.87KK1279 pKa = 11.33NEE1281 pKa = 4.03NEE1283 pKa = 4.5NYY1285 pKa = 7.42TARR1288 pKa = 11.84HH1289 pKa = 5.42EE1290 pKa = 4.43CSFVQAGDD1298 pKa = 3.21KK1299 pKa = 10.58FYY1301 pKa = 11.44LLGGRR1306 pKa = 11.84EE1307 pKa = 3.93NSKK1310 pKa = 8.31TLDD1313 pKa = 3.41VYY1315 pKa = 11.5NYY1317 pKa = 9.34TSNSWNSLVDD1327 pKa = 3.78SAPMEE1332 pKa = 4.47FNHH1335 pKa = 6.4FQATEE1340 pKa = 3.94YY1341 pKa = 10.59QGLIWVIGAFQTNSFPNEE1359 pKa = 3.82TPADD1363 pKa = 4.47FIWAFDD1369 pKa = 3.69PVAQQWIQGPEE1380 pKa = 3.73IPSGRR1385 pKa = 11.84RR1386 pKa = 11.84RR1387 pKa = 11.84GSAGLVVHH1395 pKa = 6.63NDD1397 pKa = 2.33KK1398 pKa = 11.02FYY1400 pKa = 10.56IVGGNTIGHH1409 pKa = 6.3NGGYY1413 pKa = 9.77VPWFDD1418 pKa = 4.53EE1419 pKa = 4.25YY1420 pKa = 11.74DD1421 pKa = 3.68PVLGTWTTLTNAPRR1435 pKa = 11.84SRR1437 pKa = 11.84DD1438 pKa = 3.32HH1439 pKa = 5.98FHH1441 pKa = 7.21AVVINNKK1448 pKa = 9.77LYY1450 pKa = 10.3AASGRR1455 pKa = 11.84LSGGDD1460 pKa = 3.13GGTFAPVIPEE1470 pKa = 3.52VDD1472 pKa = 3.49VYY1474 pKa = 11.66DD1475 pKa = 4.8FSTNSWSSLPSDD1487 pKa = 3.72QNIPTPRR1494 pKa = 11.84AGAIVANFNNKK1505 pKa = 8.84LVIAGGEE1512 pKa = 3.96VSTNVNALDD1521 pKa = 3.14ITEE1524 pKa = 4.94IYY1526 pKa = 10.52DD1527 pKa = 4.14PLDD1530 pKa = 3.57QSWSTGASLIHH1541 pKa = 6.57PRR1543 pKa = 11.84HH1544 pKa = 5.0GTQGIVSGNGIFVVAGSPVMGGGKK1568 pKa = 8.8QKK1570 pKa = 10.94NMEE1573 pKa = 4.52FYY1575 pKa = 10.98GSDD1578 pKa = 3.64SPTGIPLDD1586 pKa = 3.76ASEE1589 pKa = 5.27LMGPTTVDD1597 pKa = 3.7FSVQNSQDD1605 pKa = 2.85ITLDD1609 pKa = 3.67LEE1611 pKa = 4.49NGNTGIFIRR1620 pKa = 11.84SVEE1623 pKa = 4.24INGHH1627 pKa = 6.6DD1628 pKa = 4.32DD1629 pKa = 3.74DD1630 pKa = 6.52DD1631 pKa = 4.98FVIQSGNLNNVLLKK1645 pKa = 10.9SNSSKK1650 pKa = 10.51IISVDD1655 pKa = 4.11FIGAGTEE1662 pKa = 3.88KK1663 pKa = 10.82NAILTIDD1670 pKa = 3.52YY1671 pKa = 8.84GANASLDD1678 pKa = 3.15ISLSYY1683 pKa = 11.19DD1684 pKa = 3.46STLSSPDD1691 pKa = 3.17EE1692 pKa = 4.34KK1693 pKa = 11.61DD1694 pKa = 3.34NISNVFTIYY1703 pKa = 10.36PNPSSEE1709 pKa = 4.05YY1710 pKa = 8.91VYY1712 pKa = 11.31VEE1714 pKa = 4.34IKK1716 pKa = 10.56SSSIDD1721 pKa = 3.27VTDD1724 pKa = 4.65FLLFDD1729 pKa = 3.15ITGRR1733 pKa = 11.84LIKK1736 pKa = 10.15KK1737 pKa = 9.48IKK1739 pKa = 10.04NDD1741 pKa = 3.16NSEE1744 pKa = 3.51QSTFKK1749 pKa = 10.83KK1750 pKa = 10.58QIDD1753 pKa = 3.69VRR1755 pKa = 11.84GLQSGLYY1762 pKa = 8.59TLYY1765 pKa = 10.17ILSKK1769 pKa = 10.52GSVVHH1774 pKa = 6.88KK1775 pKa = 10.7ARR1777 pKa = 11.84LVINNN1782 pKa = 3.78
Molecular weight: 190.99 kDa
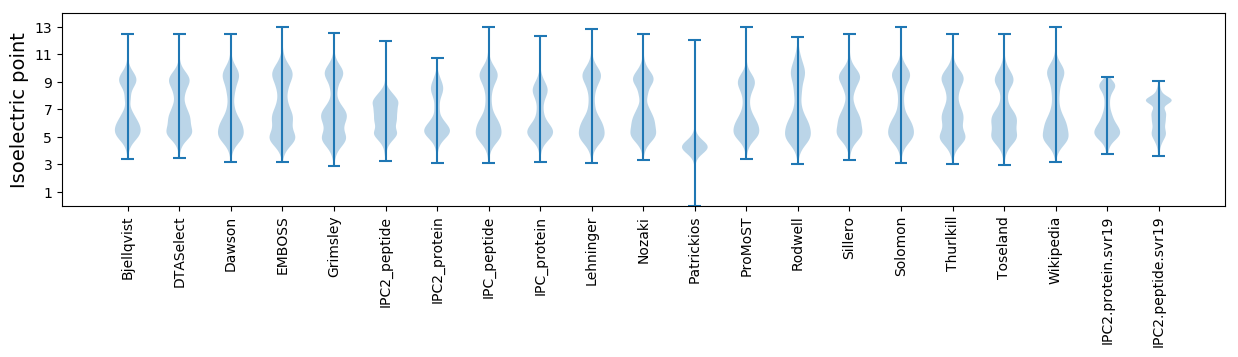
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N3HN48|A0A2N3HN48_9FLAO Uncharacterized protein OS=Confluentibacter flavum OX=1909700 GN=CSW08_04105 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1212577 |
38 |
2585 |
350.6 |
39.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.072 ± 0.037 | 0.772 ± 0.013 |
5.725 ± 0.033 | 6.317 ± 0.037 |
5.277 ± 0.032 | 6.419 ± 0.043 |
1.86 ± 0.022 | 8.324 ± 0.043 |
7.737 ± 0.048 | 9.27 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.239 ± 0.021 | 6.499 ± 0.047 |
3.417 ± 0.023 | 3.196 ± 0.025 |
3.257 ± 0.025 | 6.528 ± 0.03 |
5.79 ± 0.034 | 6.061 ± 0.031 |
1.12 ± 0.018 | 4.12 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
