
TTV-like mini virus
Taxonomy: Viruses; Anelloviridae; Betatorquevirus; unclassified Betatorquevirus
Average proteome isoelectric point is 7.36
Get precalculated fractions of proteins
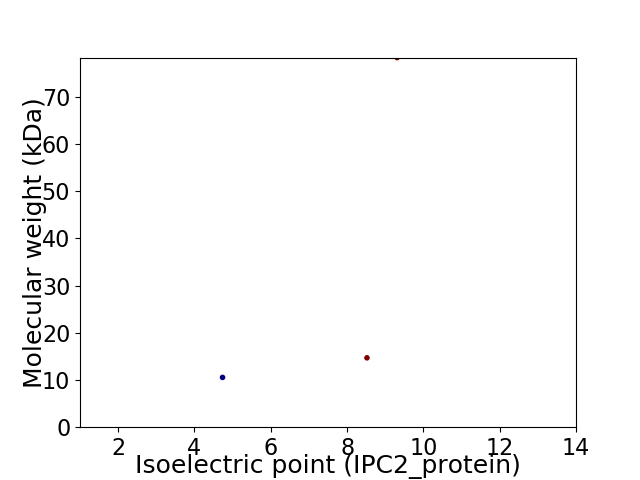
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V5NT04|V5NT04_9VIRU Uncharacterized protein OS=TTV-like mini virus OX=93678 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.62SMPFSTKK8 pKa = 9.04KK9 pKa = 10.28QRR11 pKa = 11.84LQLCNLVTGIHH22 pKa = 7.6DD23 pKa = 3.72ITCNCYY29 pKa = 10.37NPLFHH34 pKa = 7.14SAQIILKK41 pKa = 9.83QLAPEE46 pKa = 4.2LKK48 pKa = 10.27KK49 pKa = 10.86EE50 pKa = 4.29EE51 pKa = 3.96KK52 pKa = 10.22HH53 pKa = 6.25QLKK56 pKa = 9.85QCLTEE61 pKa = 3.87EE62 pKa = 4.39TTTKK66 pKa = 10.68EE67 pKa = 3.96EE68 pKa = 4.07EE69 pKa = 4.37DD70 pKa = 3.55VGFTTGDD77 pKa = 3.46LEE79 pKa = 5.31ALFAEE84 pKa = 4.82NGDD87 pKa = 3.96DD88 pKa = 4.11TEE90 pKa = 4.61DD91 pKa = 3.47AAGG94 pKa = 3.49
MM1 pKa = 7.62SMPFSTKK8 pKa = 9.04KK9 pKa = 10.28QRR11 pKa = 11.84LQLCNLVTGIHH22 pKa = 7.6DD23 pKa = 3.72ITCNCYY29 pKa = 10.37NPLFHH34 pKa = 7.14SAQIILKK41 pKa = 9.83QLAPEE46 pKa = 4.2LKK48 pKa = 10.27KK49 pKa = 10.86EE50 pKa = 4.29EE51 pKa = 3.96KK52 pKa = 10.22HH53 pKa = 6.25QLKK56 pKa = 9.85QCLTEE61 pKa = 3.87EE62 pKa = 4.39TTTKK66 pKa = 10.68EE67 pKa = 3.96EE68 pKa = 4.07EE69 pKa = 4.37DD70 pKa = 3.55VGFTTGDD77 pKa = 3.46LEE79 pKa = 5.31ALFAEE84 pKa = 4.82NGDD87 pKa = 3.96DD88 pKa = 4.11TEE90 pKa = 4.61DD91 pKa = 3.47AAGG94 pKa = 3.49
Molecular weight: 10.54 kDa
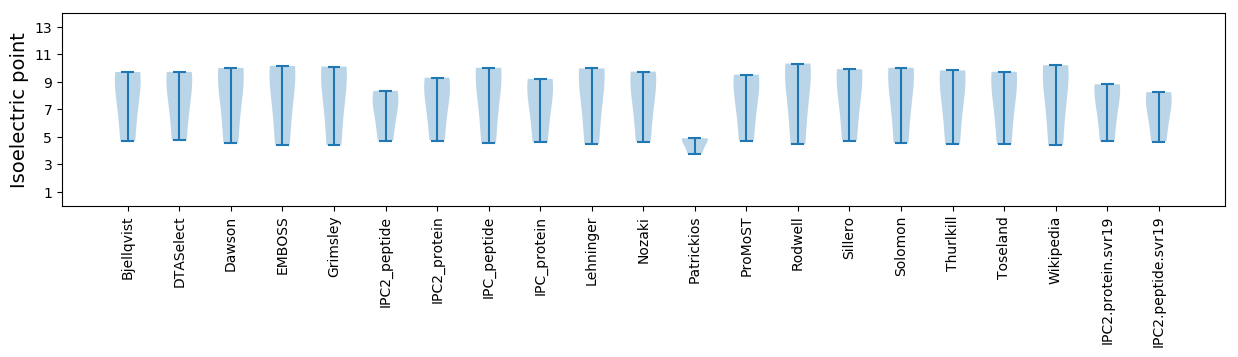
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V5NSF8|V5NSF8_9VIRU Uncharacterized protein OS=TTV-like mini virus OX=93678 GN=ORF2 PE=4 SV=1
MM1 pKa = 8.07PYY3 pKa = 10.06RR4 pKa = 11.84RR5 pKa = 11.84NYY7 pKa = 6.8YY8 pKa = 7.33QRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84WIYY17 pKa = 8.6YY18 pKa = 8.77RR19 pKa = 11.84RR20 pKa = 11.84PRR22 pKa = 11.84GPFRR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 5.31WRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 7.83RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84WVRR39 pKa = 11.84KK40 pKa = 9.13KK41 pKa = 10.29KK42 pKa = 10.85LPFLRR47 pKa = 11.84LKK49 pKa = 10.26EE50 pKa = 4.07YY51 pKa = 10.53QPSNIRR57 pKa = 11.84KK58 pKa = 9.41CKK60 pKa = 9.04IQGLIPLYY68 pKa = 9.67WGTPEE73 pKa = 5.06RR74 pKa = 11.84FVNNYY79 pKa = 10.12DD80 pKa = 3.62SYY82 pKa = 11.92EE83 pKa = 4.11MTTAPPKK90 pKa = 10.47LPSGGLFTIKK100 pKa = 10.45NFSLQSLYY108 pKa = 11.14SEE110 pKa = 3.8NRR112 pKa = 11.84YY113 pKa = 8.87IRR115 pKa = 11.84NTWSHH120 pKa = 6.23TNTDD124 pKa = 4.02LPLVRR129 pKa = 11.84YY130 pKa = 6.99TGCKK134 pKa = 8.64FKK136 pKa = 10.3IYY138 pKa = 10.12KK139 pKa = 9.63SEE141 pKa = 4.07HH142 pKa = 6.02VDD144 pKa = 3.92TILTFDD150 pKa = 4.02NNLPLASNQDD160 pKa = 3.98MYY162 pKa = 11.92LSMQPSVHH170 pKa = 7.67SMIKK174 pKa = 9.97HH175 pKa = 6.14KK176 pKa = 10.52IIIPRR181 pKa = 11.84LTKK184 pKa = 10.42GYY186 pKa = 10.21NKK188 pKa = 10.12KK189 pKa = 9.97PYY191 pKa = 8.45KK192 pKa = 9.31TIHH195 pKa = 6.27IKK197 pKa = 10.08PPKK200 pKa = 9.66PMINKK205 pKa = 9.03WYY207 pKa = 8.65FQQDD211 pKa = 2.66ITTIPLCQIRR221 pKa = 11.84ASACSLNEE229 pKa = 3.24WYY231 pKa = 9.91INYY234 pKa = 9.8KK235 pKa = 10.29SVSTTSTINFLNPGCITNTYY255 pKa = 8.43FNKK258 pKa = 10.27YY259 pKa = 10.1YY260 pKa = 9.53STGWYY265 pKa = 9.36ARR267 pKa = 11.84KK268 pKa = 9.56LDD270 pKa = 4.21SGTKK274 pKa = 9.44IYY276 pKa = 10.72LISFKK281 pKa = 11.02NNEE284 pKa = 3.48QHH286 pKa = 4.99TWEE289 pKa = 4.12NTVFLGNTLDD299 pKa = 3.76NQEE302 pKa = 3.98GKK304 pKa = 8.1TPKK307 pKa = 10.27EE308 pKa = 3.77INLKK312 pKa = 10.53SKK314 pKa = 10.86GSTTQDD320 pKa = 2.09IMQXYY325 pKa = 9.13GKK327 pKa = 8.94QQWGNPFYY335 pKa = 11.23KK336 pKa = 10.46NYY338 pKa = 10.72LNNTWKK344 pKa = 10.66VYY346 pKa = 9.68FFNEE350 pKa = 3.86EE351 pKa = 4.13LFTFLDD357 pKa = 3.97QYY359 pKa = 10.34RR360 pKa = 11.84QHH362 pKa = 7.38PNTPIPVTYY371 pKa = 8.99ITEE374 pKa = 4.28TQLTNAIRR382 pKa = 11.84YY383 pKa = 8.17NPLSDD388 pKa = 3.4EE389 pKa = 4.54GKK391 pKa = 8.76DD392 pKa = 3.08TKK394 pKa = 10.71IYY396 pKa = 9.85FKK398 pKa = 9.8PTNQPNEE405 pKa = 4.26DD406 pKa = 3.65WDD408 pKa = 4.17PPQNPNLMAVNLPLWILTFGFVDD431 pKa = 3.17YY432 pKa = 10.68HH433 pKa = 7.14KK434 pKa = 10.82KK435 pKa = 10.37LKK437 pKa = 9.19QVKK440 pKa = 10.31NIDD443 pKa = 3.45TEE445 pKa = 4.32QILTIQAKK453 pKa = 10.35LDD455 pKa = 3.61GPITKK460 pKa = 9.56VYY462 pKa = 10.18PLIDD466 pKa = 3.39SDD468 pKa = 4.73FIAGKK473 pKa = 9.96SPYY476 pKa = 9.5EE477 pKa = 4.07QVPDD481 pKa = 4.01PLDD484 pKa = 3.44YY485 pKa = 11.34NRR487 pKa = 11.84WHH489 pKa = 7.26LSFQFQQITTNNIGLSGPGSVKK511 pKa = 10.62SPTLQAVQAKK521 pKa = 9.01CHH523 pKa = 4.15YY524 pKa = 9.17TFYY527 pKa = 10.75FKK529 pKa = 10.24WGGNPPPMQPITDD542 pKa = 3.82PTKK545 pKa = 9.62QPHH548 pKa = 6.09YY549 pKa = 9.77NLPFNITKK557 pKa = 7.51TTSLQNPATSPAYY570 pKa = 10.17SLYY573 pKa = 10.06TFDD576 pKa = 3.69YY577 pKa = 10.74RR578 pKa = 11.84RR579 pKa = 11.84GXLTTKK585 pKa = 10.54AIEE588 pKa = 4.13RR589 pKa = 11.84LQKK592 pKa = 10.6DD593 pKa = 4.0FPLKK597 pKa = 9.81KK598 pKa = 9.27TFITDD603 pKa = 3.32SEE605 pKa = 4.63RR606 pKa = 11.84FQPPLQTQQEE616 pKa = 4.63TTSEE620 pKa = 4.15SSSEE624 pKa = 4.04EE625 pKa = 3.88EE626 pKa = 3.99EE627 pKa = 4.55EE628 pKa = 4.99EE629 pKa = 5.3ISTLLNKK636 pKa = 10.23LQRR639 pKa = 11.84QRR641 pKa = 11.84HH642 pKa = 4.53KK643 pKa = 10.83QKK645 pKa = 10.1QLKK648 pKa = 9.75YY649 pKa = 10.61LILKK653 pKa = 9.24NLGYY657 pKa = 10.35LPKK660 pKa = 10.71LEE662 pKa = 4.13
MM1 pKa = 8.07PYY3 pKa = 10.06RR4 pKa = 11.84RR5 pKa = 11.84NYY7 pKa = 6.8YY8 pKa = 7.33QRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84WIYY17 pKa = 8.6YY18 pKa = 8.77RR19 pKa = 11.84RR20 pKa = 11.84PRR22 pKa = 11.84GPFRR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 5.31WRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 7.83RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84WVRR39 pKa = 11.84KK40 pKa = 9.13KK41 pKa = 10.29KK42 pKa = 10.85LPFLRR47 pKa = 11.84LKK49 pKa = 10.26EE50 pKa = 4.07YY51 pKa = 10.53QPSNIRR57 pKa = 11.84KK58 pKa = 9.41CKK60 pKa = 9.04IQGLIPLYY68 pKa = 9.67WGTPEE73 pKa = 5.06RR74 pKa = 11.84FVNNYY79 pKa = 10.12DD80 pKa = 3.62SYY82 pKa = 11.92EE83 pKa = 4.11MTTAPPKK90 pKa = 10.47LPSGGLFTIKK100 pKa = 10.45NFSLQSLYY108 pKa = 11.14SEE110 pKa = 3.8NRR112 pKa = 11.84YY113 pKa = 8.87IRR115 pKa = 11.84NTWSHH120 pKa = 6.23TNTDD124 pKa = 4.02LPLVRR129 pKa = 11.84YY130 pKa = 6.99TGCKK134 pKa = 8.64FKK136 pKa = 10.3IYY138 pKa = 10.12KK139 pKa = 9.63SEE141 pKa = 4.07HH142 pKa = 6.02VDD144 pKa = 3.92TILTFDD150 pKa = 4.02NNLPLASNQDD160 pKa = 3.98MYY162 pKa = 11.92LSMQPSVHH170 pKa = 7.67SMIKK174 pKa = 9.97HH175 pKa = 6.14KK176 pKa = 10.52IIIPRR181 pKa = 11.84LTKK184 pKa = 10.42GYY186 pKa = 10.21NKK188 pKa = 10.12KK189 pKa = 9.97PYY191 pKa = 8.45KK192 pKa = 9.31TIHH195 pKa = 6.27IKK197 pKa = 10.08PPKK200 pKa = 9.66PMINKK205 pKa = 9.03WYY207 pKa = 8.65FQQDD211 pKa = 2.66ITTIPLCQIRR221 pKa = 11.84ASACSLNEE229 pKa = 3.24WYY231 pKa = 9.91INYY234 pKa = 9.8KK235 pKa = 10.29SVSTTSTINFLNPGCITNTYY255 pKa = 8.43FNKK258 pKa = 10.27YY259 pKa = 10.1YY260 pKa = 9.53STGWYY265 pKa = 9.36ARR267 pKa = 11.84KK268 pKa = 9.56LDD270 pKa = 4.21SGTKK274 pKa = 9.44IYY276 pKa = 10.72LISFKK281 pKa = 11.02NNEE284 pKa = 3.48QHH286 pKa = 4.99TWEE289 pKa = 4.12NTVFLGNTLDD299 pKa = 3.76NQEE302 pKa = 3.98GKK304 pKa = 8.1TPKK307 pKa = 10.27EE308 pKa = 3.77INLKK312 pKa = 10.53SKK314 pKa = 10.86GSTTQDD320 pKa = 2.09IMQXYY325 pKa = 9.13GKK327 pKa = 8.94QQWGNPFYY335 pKa = 11.23KK336 pKa = 10.46NYY338 pKa = 10.72LNNTWKK344 pKa = 10.66VYY346 pKa = 9.68FFNEE350 pKa = 3.86EE351 pKa = 4.13LFTFLDD357 pKa = 3.97QYY359 pKa = 10.34RR360 pKa = 11.84QHH362 pKa = 7.38PNTPIPVTYY371 pKa = 8.99ITEE374 pKa = 4.28TQLTNAIRR382 pKa = 11.84YY383 pKa = 8.17NPLSDD388 pKa = 3.4EE389 pKa = 4.54GKK391 pKa = 8.76DD392 pKa = 3.08TKK394 pKa = 10.71IYY396 pKa = 9.85FKK398 pKa = 9.8PTNQPNEE405 pKa = 4.26DD406 pKa = 3.65WDD408 pKa = 4.17PPQNPNLMAVNLPLWILTFGFVDD431 pKa = 3.17YY432 pKa = 10.68HH433 pKa = 7.14KK434 pKa = 10.82KK435 pKa = 10.37LKK437 pKa = 9.19QVKK440 pKa = 10.31NIDD443 pKa = 3.45TEE445 pKa = 4.32QILTIQAKK453 pKa = 10.35LDD455 pKa = 3.61GPITKK460 pKa = 9.56VYY462 pKa = 10.18PLIDD466 pKa = 3.39SDD468 pKa = 4.73FIAGKK473 pKa = 9.96SPYY476 pKa = 9.5EE477 pKa = 4.07QVPDD481 pKa = 4.01PLDD484 pKa = 3.44YY485 pKa = 11.34NRR487 pKa = 11.84WHH489 pKa = 7.26LSFQFQQITTNNIGLSGPGSVKK511 pKa = 10.62SPTLQAVQAKK521 pKa = 9.01CHH523 pKa = 4.15YY524 pKa = 9.17TFYY527 pKa = 10.75FKK529 pKa = 10.24WGGNPPPMQPITDD542 pKa = 3.82PTKK545 pKa = 9.62QPHH548 pKa = 6.09YY549 pKa = 9.77NLPFNITKK557 pKa = 7.51TTSLQNPATSPAYY570 pKa = 10.17SLYY573 pKa = 10.06TFDD576 pKa = 3.69YY577 pKa = 10.74RR578 pKa = 11.84RR579 pKa = 11.84GXLTTKK585 pKa = 10.54AIEE588 pKa = 4.13RR589 pKa = 11.84LQKK592 pKa = 10.6DD593 pKa = 4.0FPLKK597 pKa = 9.81KK598 pKa = 9.27TFITDD603 pKa = 3.32SEE605 pKa = 4.63RR606 pKa = 11.84FQPPLQTQQEE616 pKa = 4.63TTSEE620 pKa = 4.15SSSEE624 pKa = 4.04EE625 pKa = 3.88EE626 pKa = 3.99EE627 pKa = 4.55EE628 pKa = 4.99EE629 pKa = 5.3ISTLLNKK636 pKa = 10.23LQRR639 pKa = 11.84QRR641 pKa = 11.84HH642 pKa = 4.53KK643 pKa = 10.83QKK645 pKa = 10.1QLKK648 pKa = 9.75YY649 pKa = 10.61LILKK653 pKa = 9.24NLGYY657 pKa = 10.35LPKK660 pKa = 10.71LEE662 pKa = 4.13
Molecular weight: 78.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
882 |
94 |
662 |
294.0 |
34.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.608 ± 1.119 | 1.134 ± 0.913 |
3.968 ± 0.704 | 5.669 ± 2.271 |
4.422 ± 0.164 | 3.741 ± 0.721 |
1.927 ± 0.367 | 6.122 ± 0.793 |
8.844 ± 0.212 | 9.524 ± 1.336 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.361 ± 0.252 | 6.463 ± 1.158 |
7.37 ± 1.222 | 6.689 ± 0.877 |
4.989 ± 1.202 | 5.669 ± 0.768 |
9.524 ± 0.902 | 2.041 ± 0.639 |
1.701 ± 0.834 | 5.896 ± 1.587 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
